Abstract
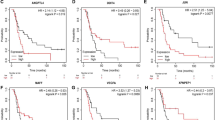
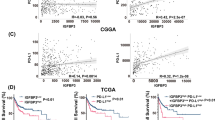
Hypoxia has been shown to contribute to tumor immunosuppressive microenvironment and is an effective prognostic indicator. This study aimed to screen prognostic hypoxia-related genes (HRGs) in glioblastoma and investigate the association between HRGs and tumor immunosuppressive microenvironment. The glioblastoma-related mRNA data were collected from TCGA, GEO, and CGGA databases. Totally 200 HRGs were obtained from the GSEA website. The prognostic HRGs were screened by univariate Cox regression analysis. Somatic mutation data of glioblastoma from TCGA was visualized using the “maftools” of R package. Immune cell infiltration proportions were calculated by CIBERSORT. The TISIDB online tool was applied to analyze the relationship between HRGs and immunoinhibitors as well as the HRG expression in different glioblastoma immune and molecular subtypes. Hub gene’s mRNA and protein levels in cell lines were determined by qRT-PCR and western blot, respectively. The effects of hub gene knockdown on cell viability and migration ability were evaluated employing CCK8 and wound healing assays. The univariate Cox regression showed that high level of FBP1 (fructose-1,6-bisphosphatase 1) was a poor prognostic biomarker, and FBP1 was mainly expressed in lymphocyte depleted immune subtype of glioblastoma. High FBP1 mRNA and protein levels have been successfully validated in vitro. The somatic mutation analysis suggested that TP53 mutation rate was the highest in the high FBP1 glioblastoma group, while EGFR mutation rate was the highest in the low FBP1 glioblastoma group. In the high FBP1 group, the infiltration proportions and types of immune cells were less, dominated by macrophages M2, and the expression of CTLA4, LAG3, TIGIT, PDL1, and PDL2 was significantly upregulated. The expression of FBP1 was positively correlated with several immunoinhibitors, such as IL-10 and TGFβ-1. In conclusion, we demonstrated that FBP1 could serve as a prognostic biomarker for glioblastoma. The immune microenvironment in the high FBP1 group might be suppressed by up-regulating immune checkpoints and immunoinhibitors.






Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available in the The Cancer Genome Atlas (TCGA) repository (https://tcga-data.nci.nih.gov/tcga/), the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/, accession number GSE4290 and GSE4412), and the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/).
References
Louis DN et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251
Shieh LT et al (2021) Epidemiologic features, survival, and prognostic factors among patients with different histologic variants of glioblastoma: analysis of a nationwide database. Front Neurol 12:659921
Jiang B et al (2021) Mortality due to primary brain tumours in China and detection rate in people with suspected symptoms: a nationally representative cross-sectional survey. World J Surg Oncol 19(1):71
Debinski W, Priebe W, Tatter SB (2017) In: DeVleeschouwer S (ed) Maximizing local access to therapeutic deliveries in glioblastoma. Part I: targeted cytotoxic therapy, in glioblastoma, Brisbane (AU)
Haemmig S et al (2014) miR-125b controls apoptosis and temozolomide resistance by targeting TNFAIP3 and NKIRAS2 in glioblastomas. Cell Death Dis 5:e1279
Arunachalam E et al (2022) HOX and PBX gene dysregulation as a therapeutic target in glioblastoma multiforme. BMC Cancer 22(1):400
Marenco-Hillembrand L et al (2020) Trends in glioblastoma: outcomes over time and type of intervention: a systematic evidence based analysis. J Neurooncol 147(2):297–307
Medikonda R et al (2021) A review of glioblastoma immunotherapy. J Neurooncol 151(1):41–53
DeCordova S et al (2020) Molecular heterogeneity and immunosuppressive microenvironment in glioblastoma. Front Immunol 11:1402
McGranahan T et al (2019) Current state of immunotherapy for treatment of glioblastoma. Curr Treat Options Oncol 20(3):24
Wang J, Chi S (2022) Characterization of the immune cell infiltration landscape and a new prognostic score in glioblastoma. J Healthc Eng 2022:4326728
Antonios JP et al (2017) Immunosuppressive tumor-infiltrating myeloid cells mediate adaptive immune resistance via a PD-1/PD-L1 mechanism in glioblastoma. Neuro Oncol 19(6):796–807
Hanihara M et al (2016) Synergistic antitumor effect with indoleamine 2,3-dioxygenase inhibition and temozolomide in a murine glioma model. J Neurosurg 124(6):1594–1601
Alam A et al (2019) SMAR1 favors immunosurveillance of cancer cells by modulating calnexin and MHC I expression. Neoplasia 21(10):945–962
Toedebusch R et al (2021) Glioma-associated microglia/macrophages augment tumorigenicity in canine astrocytoma, a naturally occurring model of human glioma. Neurooncol Adv 3(1) p. vdab062
Kurdi M et al (2021) The relationship between CD204 (M2)-polarized tumour-associated macrophages (TAMs), tumour-infiltrating lymphocytes (TILs), and microglial activation in glioblastoma microenvironment: a novel immune checkpoint receptor target. Discov Oncol 12(1):28
Trikha P, Carson WE 3rd (2014) Signaling pathways involved in MDSC regulation. Biochim Biophys Acta 1846(1):55–65
Gabrilovich DI, Nagaraj S (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9(3):162–174
Guo X et al (2018) Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten pathways. Oncogene 37(31):4239–4259
Wei J et al (2011) Hypoxia potentiates glioma-mediated immunosuppression. PLoS One 6(1):e16195
Liu J et al (2022) Carbonic anhydrase IX-targeted H-APBC nanosystem combined with phototherapy facilitates the efficacy of PI3K/mTOR inhibitor and resists HIF-1alpha-dependent tumor hypoxia adaptation. J Nanobiotechnology 20(1):187
Chambers AM, Matosevic S (2019) Immunometabolic dysfunction of natural killer cells mediated by the hypoxia-CD73 axis in solid tumors. Front Mol Biosci 6:60
Terry S, Buart S, Chouaib S (2017) Hypoxic stress-induced tumor and immune plasticity, suppression, and impact on tumor heterogeneity. Front Immunol 8:1625
He X et al (2021) Hypoxia-related gene-based signature can evaluate the tumor immune microenvironment and predict the prognosis of colon adenocarcinoma patients. Int J Gen Med 14:9853–9862
Chen X et al (2021) A hypoxia signature for predicting prognosis and tumor immune microenvironment in adrenocortical carcinoma. J Oncol 2021:2298973
Newman AM et al (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5):453–457
Yan C et al (2022) Comprehensive analysis of the immune and prognostic implication of TRIM8 in breast cancer. Front Genet 13:835540
Liu ZH et al (2015) Far upstream element-binding protein 1 is a prognostic biomarker and promotes nasopharyngeal carcinoma progression. Cell Death Dis 6(10):e1920
Barsoum IB et al (2014) Mechanisms of hypoxia-mediated immune escape in cancer. Cancer Res 74(24):7185–7190
Wang X et al (2021) Hypoxia-induced myeloid derived growth factor promotes hepatocellular carcinoma progression through remodeling tumor microenvironment. Theranostics 11(1):209–221
Yang Q et al (2020) The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B 10(11):2156–2170
Chan TA et al (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30(1):44–56
Lee EQ (2021) Immune checkpoint inhibitors in GBM. J Neurooncol 155(1):1–11
Takahashi T et al (2022) Predictive value of EGFR mutation in non-small-cell lung cancer patients treated with platinum doublet postoperative chemotherapy. Cancer Sci 113(1):287–296
Li H et al (2021) FBP1 regulates proliferation, metastasis, and chemoresistance by participating in C-MYC/STAT3 signaling axis in ovarian cancer. Oncogene 40(40):5938–5949
Son B et al (2020) Decreased FBP1 expression rewires metabolic processes affecting aggressiveness of glioblastoma. Oncogene 39(1):36–49
Chen Z et al (2023) GBE1 promotes glioma progression by enhancing aerobic glycolysis through inhibition of FBP1. Cancers (Basel) 15(5)
Yang C et al (2020) FBP1 binds to the bromodomain of BRD4 to inhibit pancreatic cancer progression. Am J Cancer Res 10(2):523–535
Zhang YP et al (2019) The involvement of FBP1 in prostate cancer cell epithelial mesenchymal transition, invasion and metastasis by regulating the MAPK signaling pathway. Cell Cycle 18(19):2432–2446
Li XR et al (2020) Knockdown of FBP1 enhances radiosensitivity in prostate cancer cells by activating autophagy. Neoplasma 67(5):982–991
Shou Y et al (2021) Determination of hypoxia signature to predict prognosis and the tumor immune microenvironment in melanoma. Mol Omics 17(2):307–316
Li CH, Chan MH, Chang YC (2022) The role of fructose 1,6-bisphosphate-mediated glycolysis/gluconeogenesis genes in cancer prognosis. Aging (Albany NY) 14(7):3233–3258
Wang J et al (2020) Adoptive cell therapy: a novel and potential immunotherapy for glioblastoma. Front Oncol 10:59
Niogret J et al (2021) Follicular helper-T cells restore CD8(+)-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J Immunother Cancer 9(6)
Yang YP et al (2021) Musashi-1 regulates MIF1-mediated M2 macrophage polarization in promoting glioblastoma progression. Cancers (Basel) 13(8)
Devalaraja S et al (2020) Tumor-derived retinoic acid regulates intratumoral monocyte differentiation to promote immune suppression. Cell 180(6):1098–1114 e16
Chin AR, Wang SE (2016) Cancer-derived extracellular vesicles: the 'soil conditioner' in breast cancer metastasis? Cancer Metastasis Rev 35(4):669–676
Frydrychowicz M et al (2017) The dual role of Treg in cancer. Scand J Immunol 86(6):436–443
Noguchi T et al (2017) Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol Res 5(2):106–117
Yuan F et al (2020) Molecular and clinical characterization of Galectin-9 in glioma through 1,027 samples. J Cell Physiol 235(5):4326–4334
Redman-Rivera LN et al (2021) Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes. Nat Commun 12(1):5184
Lin S et al (2019) Intrinsic adriamycin resistance in p53-mutated breast cancer is related to the miR-30c/FANCF/REV1-mediated DNA damage response. Cell Death Dis 10(9):666
Shi C et al (2021) A TP53 mutation model for the prediction of prognosis and therapeutic responses in head and neck squamous cell carcinoma. BMC Cancer 21(1):1035
Tsubata Y, Tanino R, Isobe T (2021) Current therapeutic strategies and prospects for EGFR mutation-positive lung cancer based on the mechanisms underlying drug resistance. Cells 10(11)
Funding
This work was supported by the Medical and Health Technology Development Plan Project of Shandong Province [grant number 2019WS308].
Author information
Authors and Affiliations
Contributions
(I) Conception and design: Hu Sun
(II) Administrative support: Bing Chen, Hu Sun, and Hui Zhang
(III) Provision of study materials or patients: Hui Zhang
(IV) Collection and assembly of data: Hao Zhao, Hu Sun, and Wei Song
(V) Data analysis and interpretation: Hao Zhao, Hu Sun, and Wei Song
(VI) Manuscript writing: all authors
(VII) Final approval of manuscript: all authors
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
Figure S1 A-B Expression levels of FBP1 under different CNV and FBP1 mutations.
ESM 2
Table S1 200 hypoxia-related genes obtained from the GSEA website.
ESM 3
Table S2 GSEA enrichment analysis revealed immune-related pathways enriched in the FBP1 high expression group.
ESM 4
Table S3 GO annotation of 281 genes in three immune-related pathways.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, H., Zhang, H., Jing, L. et al. FBP1 is a potential prognostic biomarker and correlated with tumor immunosuppressive microenvironment in glioblastoma. Neurosurg Rev 46, 187 (2023). https://doi.org/10.1007/s10143-023-02097-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10143-023-02097-y