Abstract
This study aims to explore novel and reliable biomarkers for predicting hepatocellular carcinoma (HCC) prognosis. Circular RNAs (circRNAs) were determined by analysis of human circRNA arrays and quantitative reverse transcription polymerase reactions. To test for an interaction between circDLG1, we used luciferase reporter assays, RNA immunoprecipitation, and fluorescence in situ hybridization assays that were employed to test the interaction between circDLG1, miR-141-3p, and WTAP. q-RT-PCR and western blot were used to evaluate the target regulation of miR-141-3p and WTAP. shRNA-mediated knockdown of circDLG1, proliferation, migration, and invasion experiment of metastasis were used to evaluate the function of circDLG. CircDLG1 rather than lining DLG1 was upregulated in HCC tissues, from HCC patients as well as HCC cell lines compared to normal controls. circDLG1 high expression in HCC patients was correlated with shorter overall survival. Knockdown of circDLG1 and miR-141-3p mimic could inhibit the tumorigenesis of HCC cells in vivo and in vitro. Importantly, we demonstrated that circDLG1 could act as a sponge of miR-141-3p to regulate the expression of WTAP, and further suppress the tumorigenesis of HCC cells. Our study reveals that circDLG1 can serve as a novel potential circulating biomarker for the detection of HCC. circDLG1 participates in the progression of HCC cells by sponging miR-141-3p with WTAP, providing new insight into the treatment of HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary liver cancer is one of the most common malignancies in the world, and its incidence ranks fifth among malignancies and second among tumor-related causes of death in China. Hepatocellular carcinoma (HCC) accounts for 85–90% of primary liver cancer cases (Chedid et al. 2017; Forner et al. 2012, 2018; Ranganathan et al. 2020). Current hepatocellular carcinoma treatments include surgical resection, radiofrequency ablation, transcatheter chemoembolization, radiation therapy, and molecularly targeted drugs. In spite of substantial improvements in therapeutic methods such as surgical intervention and tailored therapy, most patients with HCC are diagnosed at an advanced stage, eliminating the chance of surgical resection (Forner et al. 2018), and even with surgical treatment, most patients have a very poor prognosis due to recurrence of disease recurrence, and the 5-year survival rate is 5 ~ 30% (Hartke et al. 2017; Jiang et al. 2017; Nishida and Kudo 2017). Chemotherapy or targeted therapy is feasible for patients who cannot undergo surgery or progress postoperatively. Sorafenib is the only drug approved by the FDA for the specific treatment of advanced hepatocellular carcinoma. Unfortunately, not all patients are sensitive to it (Gao et al. 2022; Llovet et al. 2008), and the clinical benefits are limited. Furthermore, there remain a lack of reliable prognostic indicators for HCC. Patients with comparable tumor stages or pathologies can have drastically variable prognosis due to individual variations (Pinter et al. 2021; Umeda et al. 2019). Therefore, it is very important to explore new and reliable biomarkers to predict the prognosis of HCC.
In circular RNA (circRNA), a type of endogenous single-stranded ncRNA, the 3ʹ and 5ʹ nucleotides are linked together to form a covalently closed continuous ring (Chen and Yang 2015; Wilusz and Sharp 2013). It is becoming increasingly clear that circRNA is critical for the emergence of several diseases, including cancer (Hatibaruah et al. 2021). From a mechanistic point of view, circRNA can play its function in different ways, and the most common interaction is with spongy miRNA (Thomson and Dinger 2016; Yan et al. 2021). A fresh circRNA called circDLG1 was found recently. It was first identified as being highly expressed in esophageal squamous cell carcinoma and contributing to the progression of the condition (Rong et al. 2018). Furthermore, previous studies have shown that circDLG1 to be a potential marker for esophageal cancer (Zhang et al. 2020). The research on circDLG1 in tumors is very limited. Only one study has demonstrated that circDLG1 controls CXCL12 through the secretion of miR-141-3p, which promotes metastasis of gastric cancer metastasis and resistance to anti-PD-1 therapy (Chen et al. 2021), but there has been little investigation into its function or mechanism in HCC.
Here, we have shown that circDLG1 expression is elevated in hepatocellular carcinoma and can encourage the growth of HCC cells both in vivo and in vitro. CircDLG1 promotes the biological progression of HCC by activating the miR-141-3p/WTAP axis. The prospect of employing circDLG1 as a new biomarker of HCC is also highlighted by the likelihood that its elevated expression could lead to resistance to sorafenib, which is achieved by promoting HCC cell glycolysis.
Materials and methods
Cell culture
A normal liver cell line (LO2) was donated for this investigation by the Cell Resource Center of the Institute of Basic Medicine, Chinese Academy of Medical Sciences, in addition to HCC cell lines (SK-Hep-1, Huh7, HepG2, HepG3B, and BEL7404) (Beijing, China). The cell lines were grown in a 37 °C cell incubator with 5% CO2, Gibco’s Roswell Park Memorial Institute 1640 (RPMI-1640) media, 10% HyClone’s fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 U/mL penicillin, all from South Logan, UT, USA.
Cell transfection
GenePharma created the circDLG1 shRNAs (sh-circDLG1-1 and sh-circDLG1-2), mimics for miR-141-3p, inhibitors for miR-141-3p, WTAP overexpression plasmid, and associated controls (Vector, miR-NC, pcDNA, and sh-NC) (Shanghai, China). Lipofectamine 3000 was used to deliver the components to HCC cells (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time polymerase chain reaction
TaqMan miRNA assays or Takara’s PrimeScript RT reagent kit were used to reverse transcribe the RNA after it had been extracted using RNAiso Plus (Takara, Dalian, China) (Applied Biosystems, Foster City, CA, USA). Then, quantitative real-time polymerase chain reaction (qRT-PCR) was conducted using Takara’s SYBR Premix DimerEraser and associated primers (Sangon, Shanghai, China). Using the 2 − ΔΔCt normalized to GAPDH or U6, the expression was calculated. Total RNA was subjected to RNase R (3 U/g; Epicentre, Madison, WI, USA) treatment for 15 min at 37 °C to assess the features of circDLG1. The presence of circDLG1 and DLG1 levels was then evaluated.
Cell counting kit-8 assay
The capacity of cells to multiply was monitored using cell counting kit-8 (CCK-8) to identify these modifications. First, 5 × 103 cells were placed into each well of 96-well plates. After the cells had been treated in line with the experimental protocols, the well-received 10 μL of CCK-8 (Sigma-Aldrich, Shanghai, China) was left for a further 2 h. The absorbance at 450 nm was then calculated.
Colony formation assay
The HCC cells were seeded onto 12-well plates for 14 days at a density of 800 cells/well after the experimental procedures. Once the colonies were visible, the culture was discontinued. In addition, colonies were counted under a microscope after being stained with crystal violet (Sangon) (Olympus, Tokyo, Japan).
Measurement of glycolysis level
The cell supernatant was taken and centrifuged to eliminate the cell fragments after the cells had been treated in accordance with the experimental conditions. According to the manufacturer’s recommendations, we used the Sigma-Aldrich lactate test kit (Sigma-Aldrich, Shanghai, China), glucose assay kit (Sigma-Aldrich), and ECAR assay kit (Sigma-Aldrich) to assess glucose uptake, lactate production, and ECAR levels.
Western blot assay
After the cells had undergone the treatments required for the experiment, the protein was contained by lysing the cells in RIPA sample buffer (Beyotime), electrophoresed on SDS-PAGE (Solarbio, Beijing, China), and blotted onto PVDF membranes (Millipore, Billerica, MA, USA). Proteins were blocked in 5% skimmed milk for 1 h, followed by overnight exposure to primary antibodies and a secondary antibody (1:5000, CAS #pr30011; Proteintech, China) for 2 h. It was possible to observe the bands using an ECL reagent.
Dual-luciferase reporter assay
To create WT-circDGL1, MUT-circDGL1, WT-WTAP 3ʹ UTR, and MUT-WTAP 3ʹ UTR, segments of wild-type (WT) or mutant (MUT) circDGL1 or WTAP 3′ UTR missing miR-141-3p binding regions were inserted into pmirGLO (Promega, Fitchburg, WI, USA). The Dual-Luc assay was used to measure the luciferase activity (Promega, Madison, WI).
RNA immunoprecipitation assay
The treatment of the cells was done in accordance with the experiment’s guidelines. RNA immunoprecipitation (RIP) buffer was used to lyse the cells, and protein A/G Sepharose beads linked with antibody IgG or Ago2 were used to retain the lysates. The total RNA in the immunoprecipitated samples was subjected to qRT-PCR to determine the amounts of circDGL1, miR-141-3p, and WTAP.
Xenograft model
The BALB/c nude mice were divided into two groups by Beijing Vital River Laboratory Animal Technology Co., Ltd. (n = 5 per group) (Beijing, China). HCC cells that had been sh-NC or sh-circDLG1 transfected (2107) were suspended in 0.2 mL PBS before being subcutaneously injected into the mice’s flanks. The tumor size was determined after a 10-day observation period using the formula (Length × Width2) × 0.5. After 35 days, the mice were put down, and the weight of the xenograft tumors was calculated. The Affiliated Cancer Hospital of Zhengzhou University’s Ethics Committee for Animal Research granted authorization for the in vivo investigation.
Tissue acquisition
In our study, 120 HCC patients from Zhengzhou University’s Affiliated Cancer Hospital were enrolled. After the ethical council of the attached cancer hospital approved the study (Approval number: 2018ct004), the tissues from the tumor and neighboring normal tissues were removed and kept at − 80 °C until needed.
Immunohistochemistry assay
Before being treated with antibodies at 4 °C for an overnight period, tissue samples were dewaxed and hydrated. The secondary antibodies were then incubated for 10 min at room temperature following a PBS wash. Following diaminobenzidine (DAB) staining, samples were counterstained with 20% hematoxylin. The anti-PCNA (Proteintech, Wuhan, China) and anti-ki67 (Proteintech) antibodies were provided by Abcam (Abcam, UK). Immunohistochemistry (IHC) grading was carried out in accordance with the staining intensity (0, negative; 1, weak; 2, moderated; 3, strong).
RNA fluorescence in situ hybridization
Genesee Biotechnology Co., Ltd. created the FITC-labeled hsa-miR-141-3p and Cy3-labeled circDGL1 probes (Guangzhou, China). Before fixation, permeabilization in PBS containing 0.5% Triton X-100, and dehydration in ethanol, HCC cells were cultured on rounded coverslips. RNA fluorescence in situ hybridization (FISH) probes were applied to cells at 37 °C for an overnight period after being diluted (1:50), denatured, and balanced. The hybridized slides were then coated with rubber cement and left in the dark for more than 20 min. The cells were then stained with DAPI-Antifade for 10 min at room temperature. A TCS SP2 AOBS laser confocal microscope was used to see the outcomes (Leica Microsystems, Germany).
Immunofluorescence
Lab-Tek chamber slides (Nunc S/A; Polylabo, Strasbourg, France) were used to plant the cells. The cells were seeded for an additional day, permeabilized for 5 min with 0.2% Triton X-100, fixed for 15 min in 3.3% paraformaldehyde, blocked for an additional hour in 5% bovine serum albumin (BSA), and then incubated with rabbit polyclonal anti-GLUT1 and anti-HK2 antibodies for an additional day at 4 °C. The next day, goat anti-rabbit IgG that had been Alexa Fluor 550 conjugated was added to the blocking solution and was left for an hour at room temperature and in the dark. The nuclei were stained for 5 min with DAPI (4ʹ,6-diamidino-2-phenylindole). Fluorescence microscopy was then used to see the cells (BX53; Olympus, Japan).
Statistical analysis
The results from three different experiments were analyzed and presented as mean SD using GraphPad Prism 7. The association between the concentrations of miR-141-3p, circDLG1, and WTAP in HCC tissues was assessed using Spearman’s correlation coefficient. Various analyses used the Student’s t-test or a one-way analysis of variance. P < 0.05 was used to determine significance.
Results
CircDLG1 is abundantly expressed in hepatocellular carcinoma, and it is associated with poor prognosis in patients
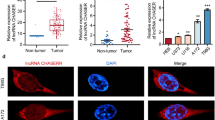
By looking at the expression of circDLG1 in tumors and nearby malignancies in the GSE97332 public data collection, we first looked at the function of circDLG1 in hepatocellular carcinoma. The results showed that tumors expressed it far more than surrounding normal tissue (Fig. 1A). As a further check on accuracy, we determined the circDLG1 expression level in fresh tissues. The outcomes matched what the database had projected would happen. Tumor expression was much higher compared to expression around cancer (Fig. 1B). In addition, we counted the overall expression trend. In hepatocellular carcinoma, 76% of patients had high expression and 24% had low expression (Fig. 1C). The expression of circDLG1 in hepatocytes and a variety of hepatoma cells in vitro was further verified. According to qRT-PCR results, Hep3B and Huh7 cell lines had the highest levels of expression, which were shown to be substantially greater in hepatoma cells than in hepatocytes (Fig. 1D). Patients with high expression also experience considerably shorter overall survival times than those with low expression (Fig. 1E). Therefore, we next applied these two cell lines for functional verification in vitro. These results show that hepatocellular carcinoma tissues and cells express more circDLG1 than healthy tissues and hepatocytes do. It provides poor prognostic information for hepatocellular cancer.
CircDLG1 is highly expressed in HCC and the prognosis of patients with high levels of expression have a poor prognosis. A Expression of the circDLG1 in HCC tissues and normal tissues in GES97332. Tumor tissue is shown in red, and normal tissue is shown in gray. B, C Expression of the circDLG1 in 120 cases of HCC tissues and normal tissues. D The circDLG1 expression in hepatocytes and five hepatocellular carcinoma cells (BEL7402, Huh7, HepG2, HepG3B, SK-hep-1, and SNU449). E Prognosis of patients with different circDLG1 expression levels. Data represent the mean ± SD of three independent experiments. All of the above experiments were performed with three biological replicates (*p < 0.05, **p < 0.01, and ***p < 0.001)
CircDLG1 is a circular structure in HCC cells
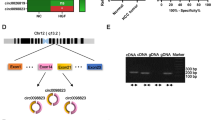
Thereafter, we digested total RNA with or without RNase R to verify that circDLG1 is a circular characteristic. These observations revealed that circDLG1 was considerably more resistant to RNase R when the MOCK group was added (Fig. 2A–E). Because circRNA has a covalently closed loop structure, it does not have the characteristics of mRNA, so we then detected the synthesis of cDNA synthesis using random primers alone. DLG1 and GAPDH can be reversed to cDNA by random primers, while circDLG1 can only form gDNA (Fig. 2G). These indicate that circDLG1 has a loop structure. It is generally established that circRNA’s function and cellular location are tightly connected. By using qRT-PCR and RNA FISH, we were able to determine circDLG1’s expression level in the cytoplasm and nucleus, respectively. It was found that about 80% of circDLG1 was located in the cytoplasm (Fig. 2C, F, and H), indicating the feasibility of its function as a miRNA sponge. These findings show that circDLG1 is a circRNA and that it is probably involved in the cytoplasm by adsorbing miRNA.
CircDLG1 is a circular structure in HCC cells. A, D The RNA levels of circDLG1 and DLG1 in HCC cells were detected by qRT-PCR. B, E qRT-PCR analysis of the circDLG1 and DLG1 expression in the HCC cells under the treatment with actinomycin D. C, F qPCR analysis of circDLG1 in the cytoplasm and nuclear of which was separated by kit. G cDNA and gDNA of HCC cells were used as the templates to amplify circDLG1, DLG1, and GAPDH with divergent primers and convergent primers, respectively. H The FISH experiment was used to detect the subcellular localization of circDLG1 in HCC cells. All of the above experiments were performed with three biological replicates (**p < 0.01 and ***p < 0.001)
CircDLG1 was shown to promote HCC cell proliferation
To further examine the role of circDLG1 in HCC, we first created a circDLG1 knockout system utilizing sh-circRNA in Hep3B and Huh7 cell lines. In HCC cells, circDLG1 expression was effectively decreased (Fig. 3A). Then, the cell viability of different groups was measured by CCK-8. According to the findings, HCC cells had lower viability due to low expression of circDLG1 (Fig. 3B and C). In order to determine the long-term alterations in the capacity of HCC cells to proliferate in the sh-circRNA group, a colony formation assay was next carried out. The circDLG1 gene deletion group had much fewer colonies than the control group (Fig. 3D and E). We also discovered changes in the mRNA and protein expression of genes related to glucose metabolism in HCC cells with varying levels of expression. The results showed that the expression of the genes GLUT1, HK2, and PKM2 involved in glucose metabolism was drastically decreased when circDLG1 was silenced (Fig. 3F and G). A similar pattern was observed in the immunofluorescence data suggesting that circDLG1 was involved in controlling the metabolism of intracellular glucose (Fig. 3H). Therefore, we further investigated how circDLG1 silencing affected cell glucose uptake and lactate generation. The results revealed that as circDLG1 expression decreased, glucose uptake and lactate production decreased (Fig. 3K and L). Glucose consumption was much lower in the knockdown group than in the control (Fig. 3I). This finding implies that circDLG1 may promote the growth of HCC cells growth by participating in glucose metabolism. As a result, we further investigated the sorafenib sensitivity of cells expressing varying amounts of circDLG1. The results showed that the circDLG1 group with low expression had a substantially lower IC50 than the group with high expression (Fig. 3J). These findings led us to make a preliminary conclusion that circDLG1 increased glycolysis in HCC cells, which in turn increased cell proliferation and made the cells more resistant to sorafenib.
CircDLG1 promoted the proliferation of HCC cells and inhibited apoptosis. A qRT-PCR analysis of the transfection efficacy of shRNA in HCC cells after 48-h transfection. B, C The proliferation status of HCC cells was determined by CCK-8 assay after circDLG1 knockdown. D, E Colony formation ability of HCC cells after circDLG1 knockdown. F The protein levels of GLUT1, PKM2, and HK2 were examined by western blot after transfected with shRNA. G The mRNA levels of GLUT1, PKM2, and HK2 were examined by RT-qPCR after transfected with shRNA. H Immunofluorescence was used to detect the expression changes of GLUT1, PKM2, and HK2 after silencing circDLG1. I, K, L Detection of ECAR (I), glucose consumption (K), and lactate production (L) in transfected cells. J The proliferation status of HCC cells was determined by CCK-8 assay after circDLG1 knockdown with or without sorafenib. All of the above experiments were performed with three biological replicates (**p < 0.01 and ***p < 0.001)
CircDLG1 promotes HCC cell proliferation in vivo
We employed a xenograft model to confirm subcutaneous carcinogenesis in vivo to further support the impact of circDLG1 on HCC. After circDLG1 was knocked down, tumor cells’ capacity to proliferate considerably diminished in comparison to the control group (Fig. 4A). The control group’s tumor volume and weight were larger than those of the knockdown group (Fig. 4B and C). Immunohistochemical detection showed that the proliferation-related indexes Ki67 and PCNA control group were significantly reduced (Fig. 4D). These in vivo results confirmed that circDLG1 was absent in vivo and inhibited tumor cell proliferation, thereby inhibiting tumor progression.
CircDLG1 promotes proliferation of HCC cells in vivo. A Images of nude mice with xenograft tumors made from transfected cells. B The line chart represented a summary of tumor volume curves. The mean standard deviation of five mice was used to represent the typical tumor volume. C The tumor mass was measured across various groups. D IHC was used to display the expression level of Ki67 and PCNA in various groups. All of the above experiments were performed with three biological replicates (**p < 0.01)
CircDLG1 has been shown to bind to miR-141-3p in HCC cells
It is generally established that circRNA’s function and cellular location are tightly connected. Our previous observations demonstrate that circDLG1 predominantly localizes to the cytoplasm, supporting the possibility that it serves as a miRNA sponge. Then, Circinteractome (https://circinteractome.nia.nih.gov/) and StarBase are used to forecast the probable targets of circDLG1, and the findings of the two databases are intersected to provide a total of six targets (Fig. 5A). The expression levels of these targets in the tumor and surrounding healthy tissues were also identified. Only has-mir-224-5p was found to be strongly expressed in the tumor, according to the data, whereas the expression of the other targets was modest. The largest expression difference was has-miR-141-3p, and the larger differences were has-mir-300 and has-mir-449a (Fig. 5B). We further tested whether these three targets bind to circDLG1. Consistent with the results, has-miR-141-3p exhibited the strongest ability to bind to circDLG1 (Fig. 5C), and has-miR-141-3p has a negative correlation with circDLG1 expression (Fig. 5D). Moreover, we predicted that miR-141-3p might be combined with circDLG1 through the online database circular RNA interactome (Circinteractome) (Fig. 5E). The binding sequence was then altered, and we discovered that ng so reduced miR-141-capacity 3p’s to bind to circDLG1 (Fig. 5F). The Argonaute2 (AGO2) protein, linked to circRNA and miRNA, forms the nucleus of the RNA-induced silencing complex (RISC), which is why we next conducted the RIP investigation. The results showed that anti-AGO2 may enrich more miR-141-3p and circDLG1 molecules than anti-IgG (Fig. 5G). These results suggest that circDLG1 may participate in HCC cells via interacting with miR-141-3p.
CircDLG1 binds to miR-141-3p in HCC cells. A The binding between circDLG1 and miR-141-3p was predicted by using Circinteractome software and StarBase, Venn graph of the intersection of two software results. B The expression levels of has-mir-224-5p, has-miR-141-3p, has-mir-300, has-mir-449a, has-mir-34a-5p, and has-mir-145-5p in the tumor and adjacent normal tissues were examined by qRT-PCR. C The interaction between circDLG1 and has-miR-141-3p, has-miR-449a, and has-miR-300 was verified by dual-luciferase reporter assay. D The expression relationship between circDLG1 and has-miR-141-3p. E CircDLG1 contained the binding sites of has-miR-141-3p. F, G The interaction between circDLG1 and miR-141-3p was verified by dual-luciferase reporter assay and RIP assay. All of the above experiments were performed with three biological replicates (*p < 0.05, **p < 0.01, and ***p < 0.001)
Targeted modulation of the expression of WTAP in HCC cells by miR-141-3p
In the 3ʹUTR of WTAP is the putative binding site for mir-141-3p. Consequently, we examined the extent to which WTAP was expressed in neighboring normal tissues and malignancy, and we found that tumors had the highest levels of WTAP expression (Fig. 6A). There was a negative correlation between miR-141-3p and WTAP expression (Fig. 6B). By using qRT-PCR and western blot, the changes in WTAP expression were discovered following the overexpression and suppression of miR-141-3p expression. The results demonstrated that miR-141-3p knockdown enhanced WTAP expression whereas miR-141-3p overexpression lowered WTAP’s mRNA and protein levels. This gave more proof that WTAP is miR-141-direct 3p’s target protein (Fig. 6C, D). In addition, we predicted that the Circinteractome online database would reveal interactions between miR-141-3p and WTAP (Fig. 6E). Then, we changed the binding sequence, and we discovered that ng so reduced mir-141-capacity 3p’s to attach to WTAP (Fig. 6F). According to these findings, miR-141-3p binds to WTAP and inhibits WTAP expression.
WTAP expression was specifically regulated by miR-141-3p in HCC cells. A GEPIA was used to verify WTAP expression in 369 HCC samples and 160 normal tissues. Normal tissue is depicted in gray, whereas tumor tissue is indicated in red. B The association between miR-141-3p levels and WTAP mRNA in HCC tissues was assessed. C A qRT-PCR experiment was used to determine the degree of WTAP mRNA expression in HCC cells. D Western blot analysis was used to determine the degree of WTAP protein expression in HCC cells. E The sequences that WTAP and miR-141-3p have in common. F Using a dual-luciferase reporter experiment, it was shown that WTAP and miR-141-3p interact. All of the above experiments were performed with three biological replicates (*p < 0.05, **p < 0.01, and ***p < 0.001)
CircDLG1 accelerated the development of HCC through the miR-141-3p/WTAP axis pathway
WTAP was overexpressed and miR-141-3p was silenced following circDLG1 silencing to demonstrate the mechanism of circDLG1 on HCC. These results demonstrated that both WTAP overexpression and miR-141-3p silencing could undo the suppression of HCC cell growth brought on by the down-regulation of circDLG1. This finding was supported by the CCK8 results (Fig. 7A and B) and colony experiment results (Fig. 7C and D). Furthermore, miR-141-3p silencing and WTAP overexpression can affect the susceptibility of HCC to sorafenib. The IC50 of sorafenib in HCC was enhanced by both overexpressing WTAP and silencing miR-141-3p after circDLG1 was silenced (Fig. 7E). We checked again to see if the metabolism of glucose powers this system. Therefore, under various settings, we were able to observe changes in the cell’s intake of glucose and the formation of lactate. The results showed that the down-regulation of glucose uptake and lactate production caused by the reduced expression of circDLG1 could be restored by up-regulating WTAP and silencing miR-141-3p (Fig. 7G and H). Results for glucose consumption revealed a similar pattern (Fig. 7F). This finding implies that circDLG1 stimulates proliferation of HCC cells through control of the miR-141-3p/WTAP axis to engage in glucose metabolism in HCC cells.
CircDLG1 promoted progression of HCC via the miR-141-3p/WTAP axis. A, B The proliferative status of HCC cells was determined by CCK-8 assay after circDLG1 knockdown with miR-141-3p inhibitor or WTAP overexpression. C, D Colony formation ability of HCC cells after circDLG1 knockdown with miR-141-3p inhibitor or WTAP overexpression. E The proliferation status of HCC cells was determined by CCK-8 assay after circDLG1 knockdown with miR-141-3p inhibitor or WTAP overexpression adding or not adding sorafenib. F–H Detection of ECAR (F), glucose consumption (G), and lactate production (H) in HCC cells after circDLG1 knockdown with miR-141-3p inhibitor or WTAP overexpression. All of the above experiments were performed with three biological replicates (*p < 0.05, **p < 0.01, and ***p < 0.001)
Discussion
With the advancement of high-throughput sequencing technology, the mechanism of disease occurrence and development has been progressively investigated, and more and more circRNAs have been identified. The role of circRNA in human disease, especially in tumor (Wei et al. 2019), cannot be ignored, even though it has been undiscovered for a long time. In recent years, there has been an increase in the study of multiple circRNA processes in tumor cells, in particular the ceRNA machinery. The ceRNA machinery is based on the idea that ceRNAs can compete with target miRNAs for binding, lowering the number of miRNAs and raising the number of mRNA targets downstream that control illness. There is a lot of evidence to suggest that ceRNA machinery plays a role in the initiation and development of some cancers, including HCC (Bai et al. 2023; Ding et al. 2023; Wang et al. 2023). In recent years, bioinformatics analysis has received increasing attention in the field of scientific research (Hu et al. 2023a, b; Zhu et al. 2022). Based on the ceRNA mechanism and bioinformatics analysis, Zhong et al. constructed a ceRNA network. Multiple gene function analysis and identification databases finally identified the association between key regulatory axes of HCC and survival prognosis, tumor-infiltrating immune cells, immune escape, pathway activity, and drug sensitivity in HCC patients, again demonstrating that the ceRNA network may promote HCC development and HCC cell invasion and progression (Zhong et al. 2023). According to a growing body of research, circRNA may affect a range of physiological and pathological processes, including apoptosis, cell cycle, cell migration, and invasion (Dong et al. 2017). CircRNA has been demonstrated to support tumor growth in a variety of mechanisms in earlier studies (Qu et al. 2017). For instance, Han et al. found that circMTO1 controls the expression of p21 via secreting miR-9, which controls the development of hepatocellular carcinoma (Han et al. 2017). Zhai et al. found that inhibition of the circIDE/miR-19b-3p/RBMS1 axis exhibited tumor-promoting activity by upregulating GPX4 to reduce iron death in hepatocellular carcinoma (Zhai et al. 2023). Hu et al. reported that exosome-derived circCCAR1 promotes CD8+ T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma (Hu et al. 2023a, b). CircFOXK2 promotes hepatocellular carcinoma progression and leads to poor clinical prognosis by modulating the Warburg effect (Zheng et al. 2023). We discovered in this work that circDLG1 is substantially expressed in hepatocellular carcinoma and that individuals with high expression of HCC had a bad prognosis. The same results were obtained in online data and our real clinical samples, which made us believe that circDLG1 plays an important role in promoting cancer in HCC. Further research found that circDLG1 regulates the glycolysis process of HCC cells.
Tumor cells, due to their rapid and unlimited proliferation, typically meet the energy requirements of cancer cells by increasing glucose uptake (Pavlova and Thompson 2016). Compared with normal cells, tumor cells have a stronger ability to absorb glucose and provide sufficient ATP for liver cancer cells through this “Warburg effect” (Zuo et al. 2021). In HCC, it has been found that multiple molecules involved in the multi-step process of glucose metabolism are abnormally expressed in HCC, which is an adverse prognostic factor of liver cancer, including GLUT1 (Amann and Hellerbrand 2009), PKM2 (Liu et al. 2017), and HK2 (DeWaal et al. 2018). Numerous variables, including circRNA, play a crucial role in controlling the expression of these genes. For instance, circ 0,058,063 upregulates the expression of GLUT1 in esophageal squamous cell carcinoma and encourages glucose absorption, which ultimately increases the course of the disease (Zheng et al. 2020). Of course, because of the unique circular structure of circRNA, it can function in different ways (Li et al. 2015; Thomson and Dinger 2016), for example, spongy miRNA, interaction with RNA binding proteins, and translation proteins. CircRNAs are more likely to play a biological role by binding miRNAs through a sponge mechanism of adsorption to regulate downstream target genes. The mechanism by which circRNA controls the expression of target genes by adsorbing miRNA to control tumor growth has also been extensively studied in the past. Circ0032821, for example, through influencing the mir-1236-3p/HMGB1 axis, stimulates the growth, migration, invasion, and glycolysis of gastric cancer cells (Chen et al. 2020). In breast cancer, Circ_0072995 upregulates SHMT2 which is mediated by the mir-149-5p (Qi et al. 2020). CircDLG1 and miR-141-3p were shown to be capable of interacting in the RIP experiment, and in this work we found that the HCC cells’ primary localization for circDLG1 is the cytoplasm. Additional evidence from a luciferase activity experiment supported miR-141-3p and circDLG1’s direct connection. These findings imply that miR-141-3p can bind to circDLG1 and control downstream protein gene expression. Our experimental findings in this paper demonstrate that circDLG1 may directly control the mRNA and protein expression of genes involved in glycolysis, including GLUT1, PKM2, and HK2, to control the degree of glucose absorption by HCC cells. This regulation can be directly reversed by miR-141-3p adsorbed by circDLG1. By adsorbing miR-141-3p, the highly expressed circDLG1 can encourage the absorption of glucose by HCC cells, boosting cell growth. This result, which is potentially a possible sorafenib resistance mechanism, was further verified in mice. The sensitivity of highly expressed circDLG1 HCC cells to sorafenib decreased. However, in this study, our research on how circDLG1 regulates the sensitivity of HCC cells to sorafenib is limited, which is our next line of research.
Since circDLG1 and miR-141-3p are not translated into proteins, we looked more closely at their genes. These studies have shown how crucial a role the WTAP protein plays in the axis of circDLG1/miR-141-3p axis. Wilms tumor 1’s partner and conserved nuclear protein is called WTAP (WT1) (Little et al. 2000). According to reports, WTAP is engaged in a variety of cellular biological processes, including alternative splicing (Haussmann et al. 2016) and cell cycle regulation (Horiuchi et al. 2006). In addition, WTAP’s function in malignancies has drawn increasing interest. According to studies, for the development and metastasis of nasopharyngeal cancer, DIAPH1-AS1 m6A must be methylated by WTAP (Li et al. 2022). WTAP is also an oncogene in pancreatic cancer and lung adenocarcinoma (Lei et al. 2022; Xu et al. 2022). WTAP promotes the growth of hepatocellular carcinoma by suppressing ETS1’s epigenetic activity in an m6A HuR-dependent manner (Chen et al. 2019), which is consistent with our research results in this paper, but whether WTAP also regulates HCC cell proliferation by regulating cell cycle–associated proteins needs further research. WTAP increases the growth of esophageal squamous cell carcinoma via lowering the expression of CPSF4 in an m6A-dependent way (Luo et al. 2022). This outcome is consistent with our research in HCC and supports the idea that miR-141-3p/WTAP, which is regulated by circDLG1, aids in the malignant proliferation of liver cancer cells.
In this study, we identified for the first time that circDLG1 is differentially expressed in hepatocellular carcinoma, and for the first time, we sub-found that circDLG1 can participate in glycolysis of HCC cells, thus mediating the proliferation and invasion and migration of HCC cells, which has not been reported in previous studies and is an important mechanism of tumor progression. However, we also have limitations in this study. First, our clinical sample size is relatively small, and there may be some bias. We need to use a larger patient cohort for further verification. In addition, We also discovered that the intracellular metabolite lactate is elevated at the same time. We regrettably did not examine whether the metabolite lactate drains into the milieu and if it alters the microenvironment using lactate, causing cell metastasis, which necessitates further investigation into the precise mechanism of circDLG1.
Conclusion
In conclusion, this study suggests that circDLG1 could be a new potential biomarker for the detection of HCC. circDLG1 is involved in mediating HCC cell glycolysis through spongy miR-141-3p with WTAP, thereby promoting HCC cell proliferation, invasion, and migration. These findings provide new insight for a more in-depth exploration of the mechanism of biological development of HCC and the search for more effective biological criteria for the treatment and prognosis of HCC.
Data availability
For data that support the findings of this study, please contact the corresponding author.
Change history
10 May 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s10142-024-01375-2
References
Amann T, Hellerbrand C (2009) GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets 13:1411–1427. https://doi.org/10.1517/14728220903307509
Bai L, Yu Z, Li Q, Hu X, Liu X, Li F, Li S, Chen Y (2023) Circ_0073228 serves as a competitive endogenous RNA to facilitate proliferation and inhibit apoptosis of hepatocellular carcinoma cells by the miR-139–5p/DNASE2 axis. J Gene Med:e3507. https://doi.org/10.1002/jgm.3507
Chedid MF, Kruel CRP, Pinto MA, Grezzana-Filho TJM, Leipnitz I, Kruel CDP, Scaffaro LA, Chedid AD (2017) Hepatocellular carcinoma: diagnosis and operative management. Arq Bras Cir Dig 30:272–278. https://doi.org/10.1590/0102-6720201700040011
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S (2019) WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer 18:127. https://doi.org/10.1186/s12943-019-1053-8
Chen L, Chi K, Xiang H, Yang Y (2020) Circ_0032821 facilitates gastric cancer cell proliferation, migration, invasion and glycolysis by regulating MiR-1236-3p/HMGB1 axis. Cancer Manag Res 12:9965–9976. https://doi.org/10.2147/cmar.s270164
Chen DL, Sheng H, Zhang DS, Jin Y, Zhao BT, Chen N, Song K, Xu RH (2021) The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol Cancer 20:166. https://doi.org/10.1186/s12943-021-01475-8
DeWaal D, Nogueira V, Terry AR, Patra KC, Jeon SM, Guzman G, Au J, Long CP, Antoniewicz MR, Hay N (2018) Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat Commun 9:446. https://doi.org/10.1038/s41467-017-02733-4
Ding N, You AB, Yang H, Hu GS, Lai CP, Liu W, Ye F (2023) A tumor suppressive-molecular axis EP300/circRERE/miR-6837-3p/MAVS activates type I interferon pathway and anti-tumor immunity to suppress colorectal cancer. Clin Cancer Res 22–3836. https://doi.org/10.1158/1078-0432.CCR-22-3836
Dong R, Ma XK, Chen LL, Yang L (2017) Increased complexity of circRNA expression during species evolution. RNA Biol 14:1064–1074. https://doi.org/10.1080/15476286.2016.1269999
Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 379:1245–1255. https://doi.org/10.1016/S0140-6736(11)61347-0
Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet 391:1301–1314. https://doi.org/10.1016/S0140-6736(18)30010-2
Gao B, Wang Y, Lu S (2022) Construction and validation of a novel signature based on epithelial-mesenchymal transition-related genes to predict prognosis and immunotherapy response in hepatocellular carcinoma by comprehensive analysis of the tumor microenvironment. Funct Integr Genomics 23:6. https://doi.org/10.1007/s10142-022-00933-w
Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (baltimore, MD) 66:1151–1164. https://doi.org/10.1002/hep.29270
Hartke J, Johnson M, Ghabril M (2017) The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol 34:153–159. https://doi.org/10.1053/j.semdp.2016.12.011
Hatibaruah A, Rahman M, Agarwala S, Singh SA, Shi J, Gupta S, Paul P (2021) Circular RNAs in cancer and diabetes. J Genet 100:21
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M (2016) m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540:301–304. https://doi.org/10.1038/nature20577
Horiuchi K, Umetani M, Minami T, Okayama H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo T, Kodama T (2006) Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci USA 103:17278–17283. https://doi.org/10.1073/pnas.0608357103
Hu F, Peng Y, Fan X, Zhang X, Jin Z (2023) Circular RNAs: implications of signaling pathways and bioinformatics in human cancer. Cancer Biol Med 20:104–128. https://doi.org/10.20892/j.issn.2095-3941.2022.0466
Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, Jiang J, Wang L, Mang Y, Gao Y, Zhang S, Ran J, Li L (2023) Exosome-derived circCCAR1 promotes CD8+ T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer 22:55. https://doi.org/10.1186/s12943-023-01759-1
Jiang JF, Lao YC, Yuan BH, Yin J, Liu X, Chen L, Zhong JH (2017) Treatment of hepatocellular carcinoma with portal vein tumor thrombus: advances and challenges. Oncotarget 8:33911–33921. https://doi.org/10.18632/oncotarget.15411
Lei J, Fan Y, Yan C, Jiamaliding Y, Tang Y, Zhou J, Huang M, Ju G, Wu J, Peng C (2022) Comprehensive analysis about prognostic and immunological role of WTAP in pan-cancer. Front Genet 13:1007696. https://doi.org/10.3389/fgene.2022.1007696
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. https://doi.org/10.1038/nsmb.2959
Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, Zhang LL, Chen F, Li YQ, Wu CF, Li F, Ma J, Liu N, Sun Y (2022) WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ 29:1137–1151. https://doi.org/10.1038/s41418-021-00905-w
Little NA, Hastie ND, Davies RC (2000) Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum Mol Genet 9:2231–2239. https://doi.org/10.1093/oxfordjournals.hmg.a018914
Liu Y, Wu H, Mei Y, Ding X, Yang X, Li C, Deng M, Gong J (2017) Clinicopathological and prognostic significance of PKM2 protein expression in cirrhotic hepatocellular carcinoma and non-cirrhotic hepatocellular carcinoma. Sci Rep 7:15294. https://doi.org/10.1038/s41598-017-14813-y
Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359:378–390. https://doi.org/10.1056/NEJMoa0708857
Luo Q, Zhan X, Kuang Y, Sun M, Dong F, Sun E, Chen B (2022) WTAP promotes oesophageal squamous cell carcinoma development by decreasing CPSF4 expression in an m(6)A-dependent manner. Med Oncol 39:231. https://doi.org/10.1007/s12032-022-01830-9
Nishida N, Kudo M (2017) Oncogenic signal and tumor microenvironment in hepatocellular carcinoma. Oncology 93(Suppl 1):160–164. https://doi.org/10.1159/000481246
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Pinter M, Scheiner B, Peck-Radosavljevic M (2021) Immunotherapy for advanced hepatocellular carcinoma: a focus on special subgroups. Gut 70:204–214. https://doi.org/10.1136/gutjnl-2020-321702
Qi C, Qin X, Zhou Z, Wang Y, Yang Q, Liao T (2020) Circ_0072995 promotes cell carcinogenesis via up-regulating miR-149-5p-mediated SHMT2 in breast cancer. Cancer Manag Res 12:11169–11181. https://doi.org/10.2147/cmar.s272274
Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H (2017) The emerging landscape of circular RNA in life processes. RNA Biol 14:992–999. https://doi.org/10.1080/15476286.2016.1220473
Ranganathan S, Lopez-Terrada D, Alaggio R (2020) Hepatoblastoma and pediatric hepatocellular carcinoma: an update. Pediatr Dev Pathol 23:79–95. https://doi.org/10.1177/1093526619875228
Rong J, Wang Q, Zhang Y, Zhu D, Sun H, Tang W, Wang R, Shi W, Cao XF (2018) Circ-DLG1 promotes the proliferation of esophageal squamous cell carcinoma. Onco Targets Ther 11:6723–6730. https://doi.org/10.2147/ott.s175826
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283. https://doi.org/10.1038/nrg.2016.20
Umeda S, Kanda M, Kodera Y (2019) Recent advances in molecular biomarkers for patients with hepatocellular carcinoma. Expert Rev Mol Diagn 19:725–738. https://doi.org/10.1080/14737159.2019.1638254
Wang J, Zheng L, Hu C, Kong D, Zhou Z, Wu B, Wu S, Fei F, Shen Y (2023) CircZFR promotes pancreatic cancer progression through a novel circRNA-miRNA-mRNA pathway and stabilizing epithelial-mesenchymal transition protein. Cell Signal 107:110661. https://doi.org/10.1016/j.cellsig.2023.110661
Wei J, Wang J, Gao X, Qi F (2019) Identification of differentially expressed circRNAs and a novel hsa_circ_0000144 that promote tumor growth in gastric cancer. Cancer Cell Int 19:268. https://doi.org/10.1186/s12935-019-0975-y
Wilusz JE, Sharp PA (2013) Molecular biology. A circuitous route to noncoding RNA. Science (new York, NY) 340:440–441. https://doi.org/10.1126/science.1238522
Xu X, Cui J, Wang H, Ma L, Zhang X, Guo W, Xue X, Wang Y, Qiu S, Tian X, Miao Y, Wu M, Yu Y, Xu Y, Wang J, Qiao Y (2022) IGF2BP3 is an essential N(6)-methyladenosine biotarget for suppressing ferroptosis in lung adenocarcinoma cells. Mater Today Bio 17:100503. https://doi.org/10.1016/j.mtbio.2022.100503
Yan LR, Ding HX, Shen SX, Lu XD, Yuan Y, Xu Q (2021) Pepsinogen C expression-related lncRNA/circRNA/mRNA profile and its co-mediated ceRNA network in gastric cancer. Funct Integr Genomics 21:605–618. https://doi.org/10.1007/s10142-021-00803-x
Zhai H, Zhong S, Wu R, Mo Z, Zheng S, Xue J, Meng H, Liu M, Chen X, Zhang G, Zheng X, Du F, Li R, Zhou B (2023) Suppressing circIDE/miR-19b-3p/RBMS1 axis exhibits promoting-tumour activity through upregulating GPX4 to diminish ferroptosis in hepatocellular carcinoma. Epigenetics 18:2192438. https://doi.org/10.1080/15592294.2023.2192438
Zhang H, Shen Y, Li Z, Ruan Y, Li T, Xiao B, Sun W (2020) The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal 34:e23049. https://doi.org/10.1002/jcla.23049
Zheng Y, Chen Y, Jiang H, Zhang H, Wang H, Xu J, Yu Z (2020) Circ_0058063 upregulates GLUT1 expression and promotes glucose-uptake in esophageal squamous-cell carcinomas. J Thorac Dis 12:925–931. https://doi.org/10.21037/jtd.2019.12.57
Zheng J, Yan X, Lu T, Song W, Li Y, Liang J, Zhang J, Cai J, Sui X, Xiao J, Chen H, Chen G, Zhang Q, Liu Y, Yang Y, Zheng K, Pan Z (2023) CircFOXK2 promotes hepatocellular carcinoma progression and leads to a poor clinical prognosis via regulating the Warburg effect. J Exp Clin Cancer Res 42:63. https://doi.org/10.1186/s13046-023-02624-1
Zhong G, Lin Y, Huang Z (2023) Identification of a novel circRNA-miRNA-mRNA regulatory axis in hepatocellular carcinoma based on bioinformatics analysis. Sci Rep 13:3728. https://doi.org/10.1038/s41598-023-30567-2
Zhu R, Gao C, Feng Q, Guan H, Wu J, Samant H, Yang F, Wang X (2022) Ferroptosis-related genes with post-transcriptional regulation mechanisms in hepatocellular carcinoma determined by bioinformatics and experimental validation. Ann Transl Med 10:1390. https://doi.org/10.21037/atm-22-5750
Zuo Q, He J, Zhang S, Wang H, Jin G, Jin H, Cheng Z, Tao X, Yu C, Li B, Yang C, Wang S, Lv Y, Zhao F, Yao M, Cong W, Wang C, Qin W (2021) PPARγ coactivator-1α suppresses metastasis of hepatocellular carcinoma by inhibiting Warburg effect by PPARγ-dependent WNT/β-catenin/pyruvate dehydrogenase kinase isozyme 1 axis. Hepatology (baltimore, MD) 73:644–660. https://doi.org/10.1002/hep.31280
Funding
This study was supported by the “Henan Province Medical Science and Technology Tackling Program (Jointly Co-construction) Project,” NO. LHGJ20190658.
Author information
Authors and Affiliations
Contributions
In terms of data analysis and article writing, QW made significant contributions to the idea and design of the work. TW made significant contributions to the analysis of the data. WY and CH made significant contributions by directing and critically reviewing the work for key intellectual substance, participating in the manuscript’s writing, responding to the referees’ comments, and giving the version to be published her seal of approval. The final text has been reviewed and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study used the patient’s tissues and was performed according to the declaration of Helsinki and was approved by the Affiliated Cancer Hospital of Zhengzhou University.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s10142-024-01375-2
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Q., Yu, W., Wang, T. et al. RETRACTED ARTICLE: Circular RNA circDLG1 contributes to HCC progression by regulating the miR-141-3p/WTAP axis. Funct Integr Genomics 23, 179 (2023). https://doi.org/10.1007/s10142-023-01096-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01096-y