Abstract
Background
Long non-coding RNA (lncRNA) ASB16 antisense RNA 1 (ASB16-AS1) is recognized as an oncogene in several cancer types, but its relation to GC is unknown. Tripartite motif containing 37 (TRIM37) has been proven to accelerate the development of gastric cancer (GC), whereas the molecular mechanism assisted ASB16-AS1 and TRIM37 in regulating GC progression remains unclear.
Methods
Differentially expressed lncRNAs in GC samples were analyzed based on Gene Expression Omnibus (GEO) data. CCK-8 and colony formation assays were applied to determine the proliferative ability of GC cells. Stem cell-like phenotype of GC cells was assessed by sphere formation assay and flow cytometry analysis. Luciferase reporter assay, RNA immunoprecipitation (RIP), pulldown, and co-immunoprecipitation (Co-IP) were performed to verify the interplay of RNA molecules.
Results
ASB16-AS1 was upregulated in GC samples according to GEO data and qRT-PCR analysis. ASB16-AS1 strengthened the proliferative ability and stem cell-like characteristics in GC cells. More importantly, ASB16-AS1 encouraged GC cell growth in vivo. Mechanistically, ASB16-AS1 strengthened TRIM37 expression by sequestering miR-3918 and miR-4676-3p. ASB16-AS1 activated NF-kappa B (NF-κB) pathway by cooperating with ATM serine/threonine kinase (ATM) to induce TRIM37 phosphorylation.
Conclusion
In summary, ASB16-AS1 exerted oncogenic functions in GC through modulating TRIM37 expression at both mRNA and protein levels.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the second commonest diagnosed cancer as well as the third leading cause of cancer-related deaths in China [1]. Transgenation, epigenetic changes, and environmental factors are major inducers promoting GC progression [2, 3]. Despite techniques in the diagnosis and treatment of GC have been improved, the 5-year survival rates in GC patients in advanced stages remain frustrating [4, 5]. Late diagnosis of GC is commonly attributed to the lack of effective diagnostic biomarkers [6]. Therefore, exploring the mechanisms underneath GC progression is valuable for the diagnosis and treatment.
Despite the fact that chemotherapy improved the prognosis of GC patients, drug resistance is a big challenge to public health. During tumor development, cancer cells can acquire stem cell-like characteristics and thereby develop drug resistance [7, 8]. Numerous factors are recognized to regulate stemness and drug resistance in GC cells. For example, SIRT1 represses stemness and drug resistance via AMPK/FOXO3 in GC [9]. SOX4 strengthens stem cell-like properties in GC [10]. Tripartite motif containing 37 (TRIM37) has been reported as an oncogene in various malignancies, including glioma [11], non-small-cell lung cancer [12] and colorectal cancer [13]. Importantly, TRIM37 promotes the resistance of pediatric osteosarcoma cells to doxorubicin, cisplatin and methotrexate [14]. Also, a study revealed the importance of TRIM37 in GC progression by verifying that TRIM37 facilitates GC cell invasion and metastasis [15]. Here, we wondered whether TRIM37 regulated the stemness and chemoresistance of GC cells.
Long noncoding RNAs (lncRNAs) are a group of non-coding RNAs with more than 200 nucleotides. Numerous works have provided evidence that lncRNAs regulate certain functional genes in GC through intricate regulatory mechanisms. For example, lncRNA UCA1 predicts unfavorable prognosis of GC and facilitates GC cell proliferation by mediating trimethylation of H3K27 in the promoters of p21 and SPRY1 [16]. OCT1-induced transcriptional activation of lncRNA DLX6-AS1 accelerates GC development via endogenously sponging miR-204-5p to up-regulate OCT1 [17]. CTC-497E21.4 boosts GC progression by serving as a competing endogenous RNA (ceRNA) to modulate miR-22/NET1 axis in an RhoA pathway-dependent way [18]. LncRNA 707 facilitates GC cell proliferation and metastasis by interacting with HuR protein to strengthen mRNA stabilities of VAV3 and F11R [19]. Functionally, lncRNAs have been documented as stemness regulators in pancreatic cancer [7], bladder cancer [8], and GC [20]. LncRNA ASB16 antisense RNA 1 (ASB16-AS1) has been identified as a crucial regulator in hepatocellular carcinoma and glioma [21, 22]. Here, we screened GEO data and found that ASB16-AS1 was upregulated in GC samples. In summary, this study was aimed at figuring out whether and how ASB16-AS1 participated in TRIM37-mediated GC progression.
Materials and methods
Tissue samples
Paired GC and adjacent non-tumor (AN) tissue samples were collected from 76 patients diagnosed with GC at Peking University Cancer Hospital & Institute between 2013 and 2018, with the written informed consents. This study recruited 46 males and 30 females, the median age was 55 years old. The division of tumors and the tumor tissue were determined in accordance with the standard surgical operation and the predetermined excision area. Stomach from each patient was cut off along the line that was over 5 cm from macroscopic tumor edge. Tissues were removed and sent to the pathology department for routine HE staining. Tissues were read by 3 senior pathologists. Tumor area was identified under the microscope and then marked. Compared with the tumor sections, the tumor and non-tumor areas were identified by observing the morphological changes under the microscope. Non-tumor tissues accounted for about 30%. Tissue samples were snap-frozen in liquid nitrogen after dissection and then preserved at − 80 °C until further analyses. Percentage of tumor cells to tumors was approximately 70% and percentage of epithelial cells to the non-tumor tissues was approximately 80–90%. This study had received the approval of the Ethics Committee of Peking University Cancer Hospital and Institute.
Cell culture and treatment
Four GC cell lines (AGS, MKN-45, HGC-27, MKN-7) and one human normal gastric epithelial cell line (GES-1) were all available from ATCC (Manassas, VA) and preserved at 37 °C with 5% CO2. RPMI-1640 (Gibco BRL, Grand Island, NY) with the supplementation of with 10% FBS (Gibco) and 1% Pen/Strep solution was used for cell culture. The anticancer agent cisplatin used to treat HGC-27 and MKN-45 cells was available from Sigma-Aldrich (St. Louis, MI). Besides, 20 μg/mL of actinomycin D (ActD) and 40 μg/mL of cycloheximide (CHX) were also acquired from Sigma-Aldrich.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNAs were isolated from GC tissues and cells using TRIzol method (Invitrogen, Carlsbad, CA) and purified using RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was generated from the total RNA (500 ng) utilizing the PrimeScript™ RT reagent kit (Takara Bio, Shiga, Japan). qRT-PCR was carried out with SYBR Green Master Mix (Takara Bio). Expressions of lncRNA and mRNA were normalized to GAPDH and that of miRNAs was normalized to U6. Relative gene expression was quantified by comparative CT method.
Cell transfection
The specific shRNAs for ASB16-AS1, RP11-475B2.1, AC010518.3 or ATM, control shRNAs, pcDNA3.1 inserted with ASB16-AS1 or TRIM37 (pcDNA3.1/ASB16-AS1 and pcDNA3.1/TRIM37), pcDNA3.1 empty vector, miR-3918 mimics/inhibitor, miR-4676-3p mimics/inhibitor, and NC mimics/inhibitor were synthesized by Genepharma (Shanghai, China). Plasmids were transfected into MKN-45 and HGC-27 cells utilizing Lipofectamine 2000 (Invitrogen) as required.
Western blot
Cells were lysed in the cold RIPA Lysis Buffer, centrifuged, subjected to electrophoresis on 10% denaturing SDS-PAGE gels, and transferred to PVDF membranes. The blots were probed with the specific primary antibodies (1: 2000) and HRP-tagged secondary antibodies (1: 5000). The visualization of blots was made using ECL detection system (Bio-Rad lab, Hercules, CA, USA). All antibodies used were produced by Abcam (Cambridge, MA, USA).
Fluorescence in situ hybridization (FISH)
The RNA FISH probes were specifically designed by Ribobio (Guangzhou, China) and utilized in line with the standard method. Cell nuclei were visualized via DAPI staining using Olympus fluorescence microscope (Olympus Corp., Tokyo, Japan).
Subcellular fractionation
Both the nuclear and cytoplasmic RNAs were isolated from the cultured GC cells (MKN-45 and HGC-27) using the PARIS™ Kit (Invitrogen). qRT-PCR was conducted to detect ASB16-AS1 expression in both fractions, with GAPDH as the cytoplasmic control and U6 as the nuclear control.
Cell counting kit 8 (CCK-8)
Cells (2 × 104) were seeded into each well of 96-well plates with or without adding cisplatin (0, 2, 4, 8 μg/mL) and incubated for 0, 24, 48, and 72 h. 10% CCK-8 solution was added for 1 h at 37 °C. The absorbance (450 nm) was evaluated utilizing microplate reader (Bio-Rad).
Colony formation assay
GC cells were added in each well of 6-well plates at a density of 500 cells per well and underwent 14-day culture for colony forming. After being fixed with 4% formaldehyde and stained with 0.5% crystal violet, colonies were counted manually.
Sphere formation assay
GC cells were seeded at 10 cells per well into the 96-well ultralow attachment plates (Corning Inc., New York, NY) containing sphere medium. After 7-days’ culture, sphere cells were counted and images were captured.
Flow cytometry analysis
The phycoerythrin (PE)-conjugated anti-CD133 antibody (BD Biosciences, San Jose, CA) was used for flow cytometry analysis. After fixation and permeabilization, cells were treated with 1% BSA for 20 min and probed with 1 μL of PE-CD133 for 1 h at 4 °C. Following the final washing, cells were examined by the flow cytometry (BD Biosciences).
RNA pull-down assay
The extracts from GC cells were acquired to mix with the biotinylated ASB16-AS1, miR-4676-3p or miR-3918 probes for 1 h. 50 μL of magnetic beads were then added for 30 min. The RNAs and proteins in the pulldown were collected, purified, and assessed by qRT-PCR and western blotting.
Luciferase reporter assay
The fragments of ASB16-AS1 or TRIM37 covering the wild-type or mutant miR-3918 or miR-4676-3p binding sites were acquired to construct the ASB16-AS1-WT/MUT and TRIM37-WT/MUT luciferase reporters using pmirGLO vectors (Promega, Madison, WI). After co-transfected with miR-3918 mimics, miR-4676-3p mimics or NC mimics for 48 h, the firefly luciferase and Renilla luciferase were both detected in Luciferase Reporter Assay System (Promega). Besides, the TRIM37 promoter was cloned into pGL3 vector (Promega) and co-transfected with ASB16-AS1 silencing transfection plasmids for further analysis.
RNA immunoprecipitation (RIP)
After lysed in the RIP lysis buffer, cell lysates were collected for immunoprecipitation with human Ago2 antibody or ATM antibody. Negative control IgG antibody was used. After incubated with the addition of magnetic beads, the specific RNAs in the binding complexes were assessed by qRT-PCR.
Northern blot
After the total cellular RNA was isolated and purified, samples were transferred onto the Hybond N+ nylon membrane, and then hybridized for 12 h with specific labeled probes targeting ASB16-AS1. GAPDH served as internal control. Finally, all membranes were exposed to phosphor screen for 48 h.
Immunofluorescence (IF)
Cells were seeded on culture slides for 24 h, rinsed in PBS and fixed for 10 min. After blocking in 5% BSA, primary anti-ATM antibody and secondary antibody (both, Abcam) were incubated with slides. The slides were finally cultured in DAPI solution and then subjected to detection with a microscope.
Co-immunoprecipitation (Co-IP)
Lysates of GC cells were suspended in IP lysis buffer for incubation with the specific antibodies or negative control IgG overnight. Then, beads were mixed with above mixtures for 2 h and the washed in IP lysis buffer, followed by the immunoprecipitated proteins analyzed via western blot.
Xenograft tumor study
The adult male BALB/C nude mice (16–20 g) obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) were preserved under SPF-grade animal lab, with the approval from the Animal Research Ethics Committee of Peking University Cancer Hospital and Institute. 5 × 106 transfected GC cells were inoculated subcutaneously to mice for 28-day to produce xenografts, with tumor volume examined every 4 days. The tumors were dissected from mice, weighed, embedded in paraffin and sectioned for analysis.
Immunohistochemistry (IHC)
Tissue samples collected from tumors in xenografts were sectioned to splices that were 4-μm thick. Then, the 4-μm paraffin-embedded sections were examined by IHC analysis using the antibodies against Ki-67, PCNA and TRIM37 (Abcam).
Statistical analyses
The results were presented as the mean ± SD, with at least three triplicates for all assays. Data analysis for this study was achieved by one-way ANOVA or student’s t test by the use of SPSS v.19.0 (SPSS Inc., Chicago, IL). Data were defined as statistically significant when P value less than 0.05.
Results
Up-regulation of ASB16-AS1 in GC tissues and cells
To identify the lncRNAs participating in GC, we analyzed GEO data (GES109476). Consequently, we obtained 474 lncRNAs up-regulated in GC tissues compared with normal samples (P < 0.05, logFC > 1). Through qRT-PCR, we further detected these 474 lncRNAs in 3 pairs of GC tissues and identified 3 most significantly up-regulated lncRNAs (ASB16-AS1, RP11-475B2.1 and AC010518.3) in GC (Fig. 1a). Next, we confirmed the high levels of the 3 lncRNAs in GC cells versus the normal gastric epithelial cell (Fig. 1b).
Up-regulation of ASB16-AS1 in GC tissues and cells. a LncRNAs upregulated in GC samples were screened out from GEO data. Heat map showed the qRT-PCR data of these 474 lncRNAs in 3 pairs of GC tissues versus adjacent normal samples. b qRT-PCR detected the levels of ASB16-AS1, RP11-475B2.1 and AC010518.3 in GC cells and normal gastric epithelial cell. c Results qRT-PCR and western blotting of TRIM37 expression in MKN-45 and HGC-27 cells with ASB16-AS1 depletion. d qRT-PCR data of ASB16-AS1 expression in adjacent non-tumor tissues (n = 76) and tumor tissues (n = 76). e qRT-PCR analysis of ASB16-AS1 expression in GC tissues at stage III-IV (n = 40) versus that at stage I-II (n = 36). f FISH image of ASB16-AS1 distribution (red) in GC cells. Scale bar: 10 μm. g Subcellular fraction assay detected percentage of ASB16-AS1 in the cytoplasm or nucleus to the whole cell. *P < 0.05, **P < 0.01
Then, we silenced ASB16-AS1, RP11-475B2.1 and AC010518.3 in MKN-45 and HGC-27 cells (Fig. S1A). Interestingly, both the levels of TRIM37 mRNA and protein were reduced only by sh-ASB16-AS1#1/2 (Fig. 1c), but not by sh-RP11-475B2.1#1/2 or sh-AC010518.3#1/2 (Fig. S1B, C). Additionally, we overexpressed ASB16-AS1 in AGS, MKN-7 and normal GES-1 cells (Fig. S1D). Results exhibited that TRIM37 level was elevated by ASB16-AS1 overexpression in AGS and MKN-7 cells but not in GES-1 cells (Fig. S1E). Therefore, we deduced that ASB16-AS1 potentially exerted functions in GC cells via regulating TRIM37.
Then, we validated that ASB16-AS1 was up-regulated in GC tissues (n = 76) versus the adjacent non-tumor tissues (n = 76) (Fig. 1d). Also, ASB16-AS1 expression was higher in tissues samples obtained from patients at III-IV (n = 40) stages than those from patients at I-II stages (n = 36) (Fig. 1e), indicating that ASB16-AS1 was potentially related to GC development. Besides, using FISH probe, we discovered that ASB6-AS1 expression was enriched in the cytoplasm (Fig. 1f), and such phenomenon was further validated by cytoplasm/nucleus fraction assay (Fig. 1g). In this regard, we speculated that ASB16-AS1 might regulate TRIM37 at post-transcriptional level.
ASB16-AS1 accelerated proliferation and strengthened stemness and cisplatin resistance of GC cells
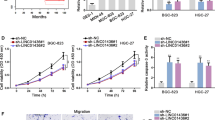
Next, in vitro functional assays were performed to detect the role of ASB6-AS1 in GC cells. CCK-8 data indicated that ASB16-AS1 depletion reduced GC cell viability (Fig. 2a). The GC cells generated less colonies under ASB6-AS1 knockdown (Fig. 2b). Then, we tested whether ASB16-AS1 regulated stem cell-like characteristics in GC cells. We observed that knocking down ASB16-AS1 declined tumor sphere formation efficiency of GC cells (Fig. 2c). According to flow cytometry analysis, CD133+ GC cells presented a decreased ratio under ASB16-AS1 deficiency (Fig. 2d). Additionally, we tested the cisplatin-resistance to determine the stemness of GC cells. CCK-8 showed that the cell viability of GC cells was gradually reduced with the treatment of increasing dose of cisplatin, and sh-ASB16-AS1#1/2 strengthened such effect (Fig. 2e). Based on CCK-8 data, cell viability was significantly reduced by cisplatin at the dose of 4 μM, and was reduced by nearly 50% at the dose of 8 μM, so we chose 5 μM for subsequent apoptosis detection. As shown in Fig. 2f, apoptosis ratio was increased by ASB16-AS1 deficiency under 5 μM cisplatin treatment, suggesting a declined resistance of ASB16-AS1-silenced GC cells to cisplatin.
Down-regulation of ASB16-AS1 hindered proliferation, stemness, and cisplatin resistance of GC cells. a Histogram diagram of CCK-8 results illustrated GC cell viability responding to ASB16-AS1 knockdown. b Pictures of colonies and the quantification of colony numbers of GC cells with ASB16-AS1 depletion. c Images of tumor spheres formed by GC cells with ASB16-AS1 silence. Sphere forming efficiency was calculated. Scale bar: 100 μm. d Flow cytometry sorted the CD133+ GC cells with ASB16-AS1 knockdown, and the CD133+ cell ratio was calculated. e CCK-8 detected the viability of GC cells with cisplatin treatment at the dose of 2, 4, and 8 μM relative to 0 μM. f Flow cytometry measured apoptosis ratio of GC cells under cisplatin treatment at 5 μM. **P < 0.01
Also, we tested the impact of ASB16-AS1 overexpression on GC cells. It was confirmed that overexpressing ASB16-AS1 in AGS and MKN-7 cells accelerated proliferation as well as colony formation (Fig. S2A, B). AGS and MKN-7 cells presented higher sphere-forming efficiency and CD133+ ratio under ASB16-AS1 overexpression (Fig. S2C, D). Cisplatin resistance in AGS and MKN-7 cells was strengthened by ASB16-AS1 overexpression (Fig. S2E, F). Additionally, we tested whether ASB16-AS1 overexpression affected normal GES-1 cells. Results showed that the proliferation, colony formation, and stem cell-like properties of GES-1 cells were not altered under ASB16-AS1 overexpression (Fig. S3A–F). Based on these findings, we suggested that ASB16-AS1 accelerated proliferation, and strengthened stemness and cisplatin resistance of GC cells.
ASB16-AS1 up-regulated TRIM37 in GC cells via interacting with miR-3918 and miR-4676-3p
Later, we investigated how ASB16-AS1 altered TRIM37 expression. We discovered that TRIM37 promoter activity was not changed under ASB16-AS1 knockdown (Fig. S4A). Also, TRIM37 mRNA was not pulled down by ASB16-AS1 biotin probe (Fig. S4B). Thus, we speculated that ASB16-AS1 post-transcriptionally regulated TRIM37 not by directly interacting with TRIM37 mRNA.
LncRNAs can post-transcriptionally alter gene expression by serving as the miRNA sponges, so we planned to explore whether ASB16-AS1 sponged certain miRNA to up-regulate TRIM37. Based on the prediction results from ENCORI database [23], 12 putative miRNAs were found to potentially bind to both lncRNA ASB16-AS1 and TRIM37 mRNA (Fig. 3a). Among the 12 miRNAs, only miR-3918, miR-4676-3p and miR-873-3p were significantly down-regulated in GC tissues (Fig. S4C), indicating that they were related to GC. Further, pull-down assay confirmed that miR-3918 and miR-4676-3p, rather than miR-873-3p, were pulled down by biotin-labeled ASB16-AS1 (Fig. 3b). It was also verified that both miR-3918 and miR-4676-3p were significantly down-regulated in GC cells (Fig. S4D). Correlation analysis based on TCGA data showed that miR-4676-3p was negatively related to ASB16-AS1, and such relationship between them was depicted based on 76 GC samples (Fig. S4E). Thus, we speculated that miR-3918 and miR-4676-3p might be involved in ASB16-AS1/TRIM37 signaling in GC.
ASB16-AS1 up-regulated TRIM37 via interacting with miR-3918 and miR-4676-3p. a ENCORI database revealed 12 miRNAs which bound both ASB16-AS1 and TRIM37. b RNA pull-down assay detected relative enrichment of miR-3918, miR-4676-3p and miR-873-3p pulled down by biotin-labeled ASB16-AS1. c The binding sites of miR-3918 or miR-4676-3p in ASB16-AS1-WT/Mut and TRIM37-WT/Mut were predicted by ENCORI. d Luciferase reporter assay examined the luciferase activity of ASB16-AS1-WT/Mut and TRIM37-WT/Mut under miR-3918 or miR-4676-3p overexpression. e RNA pull-down assay detected the relative enrichment of ASB16-AS1 and TRIM37 pulled down by biotin-labeled miR-3918 and miR-4676-3p. f RIP assay measured the enrichment of ASB16-AS1, miR-3918, miR-4676-3p and TRIM37 in the precipitates conjugated to anti-Ago2 relative to anti-IgG. g qRT-PCR, western blot and northern blot examined effects of miR-3918 and miR-4676-3p overexpression on the expression levels of TRIM37 and ASB16-AS1. h qRT-PCR and western blot detected TRIM37 expression in five different groups. i Western blot analysis of NF-κB pathway-related proteins in different groups. *P < 0.05, **P < 0.01, n.s. suggested no significance
Thereafter, we aimed to determine the binding of miR-3918 and miR-4676-3p to ASB16-AS1 and TRIM37 in GC. The putative binding sites of miR-4676-3p/miR-3918 in ASB16-AS1 and TRIM37 were predicted from ENCORI database, and the binding sites were mutated to generate ASB16-AS1-Mut and TRIM37-Mut reporters (Fig. 3c). After up-regulating miR-4676-3p and miR-3918 in GC cells (Fig. S4F), the luciferase activity of ASB16-AS1-WT and TRIM37-WT decreased, while that of ASB16-AS1-Mut and TRIM37-Mut were not impacted (Fig. 3d). Also, pull down assay revealed that ASB16-AS1 and TRIM37 could be pulled down by biotin-labeled miR-4676-3p and miR-3918 (Fig. 3e). Further, ASB16-AS1, miR-3918, miR-4676-3p and TRIM37 were co-enriched in Ago2 RIP group (Fig. 3f). These data confirmed that miR-4676-3p and miR-3918 bound to lncRNA ASB16-AS1 and TRIM37 mRNA in Ago2-assembled RNA-induced silencing complexes (RISCs). Subsequently, we detected the influence of miR-4676-3p and miR-3918 up-regulation on ASB16-AS1 and TRIM37 expression. The results revealed that miR-4676-3p and miR-3918 up-regulation lessened TRIM37 expression but had no influences on ASB16-AS1 expression (Fig. 3g). Moreover, ASB16-AS1 depletion reduced both mRNA and protein levels of TRIM37, and such effect was partly reversed by miR-4676-3p inhibitor or miR-3918 inhibitor but fully achieved by the co-inhibition of miR-4676-3p and miR-3918 (Fig. 3h). Then, NF-κB pathway was analyzed since TRIM37 has been reported to activate NF-κB pathway [12], and this pathway is supported to accelerate GC development and stemness [24, 25]. It was revealed that miR-4676-3p inhibitor or miR-3918 inhibitor recovered the levels of p-IKK, p-IκB and Nuclear p65, and such recovery was further strengthened by co-transfecting miR-4676-3p inhibitor and miR-3918 inhibitor (Fig. 3i), indicating that ASB16-AS1 might have an alternative way to regulate NF-κB pathway. In summary, ASB16-AS1 up-regulated TRIM37 via interacting with miR-3918 and miR-4676-3p to activate NF-κB pathway in GC cells.
ASB16-AS1 synergistically worked with ATM to modulate TRIM37
The full restoration of TRIM37 level caused by miR-3918/miR-4676-3p co-inhibition in ASB16-AS1-silenced GC cells indicated that miR-3918 and miR-4676-3p might be the main pathway for ASB16-AS1 to regulate TRIM37 expression. To further confirm this indication, we tested the mRNA and protein stability of TRIM37 under ASB16-AS1 knockdown. Intriguingly, degradation of TRIM37 mRNA induced by ActD treatment was not altered by ASB16-AS1 deficiency (Fig. S5A). In addition, ASB16-AS1 could not affect the protein stability of TRIM37 under CHX treatment (Fig. S5B). We previously found that NF-κB pathway was not fully recovered by miR-3918/miR-4676-3p co-inhibition, so we wondered whether ASB16-AS1 could regulate TRIM37 through other mechanisms. Pulldown and mass spectrometry revealed that ATM was a protein that could interact with ASB16-AS1 (Fig. 4a). A previous study uncovered that ATM induced the phosphorylation of TRIM37 protein in the cytoplasm and promoted nuclear translocation of TRIM37 [26]. Therefore, we speculated that ASB16-AS1 might regulate TRIM37 phosphorylation through ATM.
ASB16-AS1 synergistically worked with ATM to modulate TRIM37 at post-translational level. a RNA pull-down silver staining and mass spectrometry identified the proteins interacted with ASB16-AS1. Results were further proven by western blot analysis. b RIP assay detected the ASB16-AS1 enrichment precipitated by anti-ATM relative to anti-IgG. c FISH/IF verified that ASB16-AS1 and ATM were co-localized in the cytoplasm of GC cells. Scale bar: 10 μm. d Western blot of TRIM37 in the IP of ATM and ATM in the IP of TRIM37 in GC cells. e Western blotting analysis of p-TRIM37 expression under ATM depletion. f, g HA-tagged TRIM37 and Flag-tagged ATM were transfected into GC cells for Co-IP assay, western blot detected HA-TRIM37 in the IP of Flag and Flag-ATM in the IP of HA under ASB16-AS1 depletion in GC cells. h qRT-PCR and western blot analyses detected the influence of ASB16-AS1 depletion on ATM expression. i qRT-PCR and northern blot detected the influence of ATM depletion on ASB16-AS1 expression. j Western blot probed HA-tagged TRIM37, Flag-tagged ATM, TRAF6 and NEMO in the IP of HA by ASB16-AS1 knockdown in GC cells. **P < 0.01, n.s. suggested no significance
We observed the enrichment of ASB16-AS1 in ATM RIP products, further revealed the interaction between ASB16-AS1 and ATM (Fig. 4b). Moreover, FISH/IF verified that ASB16-AS1 and ATM were co-localized in the cytoplasm in GC cells (Fig. 4c). Besides, Co-IP assay testified the interaction between ATM and TRIM47 (Fig. 4d). More importantly, the level of p-TRIM47 was significantly reduced after ASB16-AS1 was knocked down (Fig. 4e). On the contrary, overexpressing ASB16-AS1 induced p-TRIM37 level in AGS and MKN-7 cells, whereas no changes were observed in normal GES-1 cells (Fig. S5C). Furthermore, we transfected the exogenous HA-tagged TRIM37 and Flag-tagged ATM to test the effect of ASB16-AS1 on ATM-TRIM37 interaction. It was observed that knockdown of ASB16-AS1 reduced Flag-ATM in the precipitates of HA and reduced HA-TRIM37 in the precipitates of Flag, indicating that silencing ASB16-AS1 reduced the interaction between ATM and TRIM37 (Fig. 4f, g). Meanwhile, we identified that ASB16-AS1 silence had no effects on mRNA and protein levels of ATM (Fig. 4h). Besides, we knocked down ATM (Fig. S5D), and identified that ATM knockdown could not influence ASB16-AS1 level (Fig. 4i). It was documented that TRIM37, TRAF6 and NEMO constituted the complex to modulate NF-κB activity [26]. Expectedly, we confirmed that ASB16-AS1 knockdown blocked the interaction of HA-TRIM37 with TRAF6 and NEMO (Fig. 4j). Based on these data, we reached the conclusion that ASB16-AS1 synergistically work with ATM to promote TRIM37 phosphorylation.
TRIM37 participated in ASB16-AS1-mediated GC cell proliferation, stemness and cisplatin resistance
Subsequently, we wondered whether TRIM37 could mediate the regulation of ASB16-AS1 on GC cell proliferation and stemness. Western blotting displayed that TRIM37 overexpression restored the levels of p-IKK, p-IκB and Nuclear p65 reduced by ASB16-AS1 depletion (Fig. 5a). Then, CCK-8 and colony formation assays revealed that TRIM37 up-regulation counteracted the prohibitive effect of ASB16-AS1 silence on GC cell proliferation (Fig. 5b, c). Moreover, TRIM37 overexpression restored sphere formation efficiency which was previously decreased by ASB16-AS1 knockdown in GC cells (Fig. 5d). TRIM37 overexpression recovered cisplatin resistance that was weakened by ASB16-AS1 knockdown in GC cells (Fig. 5e, f). Thus, we concluded that TRIM37 was a responsible mediator for ASB16-AS1 to influence proliferation, stemness and cisplatin resistance in GC cells.
TRIM37 participated in ASB16-AS1-mediated GC cell proliferation, stemness and cisplatin resistance. a Western blot analysis of NF-κB pathway-related proteins in sh-NC group, sh-ASB16-AS1#1 group, sh-ASB16-AS1#1+pcDNA3.1 group and sh-ASB16-AS1#1+pcDNA3.1/TRIM37 group. b, c GC cell viability and proliferation were detected in different groups through CCK-8 and colony formation assays. d Pictures of spheres were taken and sphere formation efficiency was calculated in GC cells of different groups. Scale bar: 100 μm. e, f CCK-8 and flow cytometry analysis detected GC cell viability and apoptosis in different groups with the treatment of cisplatin. **P < 0.01
ASB16-AS1 facilitated GC tumor growth in vivo through TRIM37
Finally, we explored the role of ASB16-AS1 in tumor growth through in vivo study. Nude mice xenograft model was established by subcutaneously injecting HGC-27 cells with different transfections. As illustrated in Fig. 6a, ASB16-AS1 knockdown inhibited tumor growth, and up-regulation of TRIM37 recovered tumor growth compared with ASB16-AS1 knockdown group. Also, ASB16-AS1 knockdown caused a decrease in tumor volume and weight, but such effect was counteracted by co-transfection of pcDNA3.1-TRIM37 (Fig. 6b, c). Besides, CD133+ cell ratio in xenografts was decreased by ASB16-AS1 silence but was restored by the co-transfection of pcDNA3.1-TRIM37 (Fig. 6d). Next, we confirmed that in these xenografts, both ASB16-AS1 and TRIM37 levels were reduced by ASB16-AS1 knockdown but overexpressing TRIM37 only recovered TRIM37 level rather than ASB16-AS1 level (Fig. 6e, f). Besides, TRIM37 overexpression recovered p-IKK, p-IκB and Nuclear p65 levels that were reduced by ASB16-AS1 depletion (Fig. 6f). Immunohistochemistry staining assay further indicated that Ki-67, PCNA and TRIM37 positivity in the in vivo tumors was decreased by ASB16-AS1 depletion but was rescued by the co-transfection of pcDNA3.1-TRIM37 (Fig. 6g). In sum, ASB16-AS1 facilitated GC tumor growth in vivo through a TRIM37-mediated way.
ASB16-AS1 facilitated GC tumor growth in vivo through TRIM37. a GC cells with indicated transfections were injected into the nude mice (5 mice in each group of 4 groups). Tumor growth curves were produced by analyzing tumor volume every 4 days until the 28th day. b, c Tumor volume and weight in different groups were determined at the 28th day. d Flow cytometry analysis revealed CD133+ cell ratio in xenografts. e qRT-PCR detected ASB16-AS1 and TRIM37 expressions in xenografts of different groups. f Western blot analysis of TRIM37 and NF-κB pathway-associated proteins in tumors of different groups. g Immunohistochemistry staining revealed the positivity of Ki-67, PCNA and TRIM37 in different groups. Scale bar: 100 μm. **P < 0.01
Discussion
GC is a prevalent malignancy worldwide. Recent years, the functional molecules have been identified in GC development by mechanistic investigations. For example, HOXA11 [27], STRA6 [28] and USP22 [29] are identified as the putative biomarkers for GC treatment. These findings unveiled the importance of recognizing novel targets for GC treatment. According to former studies, TRIM37 promotes drug resistance, proliferation, and metastasis in various cancers, including GC [11, 12, 14, 15]. However, our current study was the first trying to explain how TRIM37 was regulated in GC. Present study started from TRIM37 and explored the upstream mechanism of TRIM37 in GC. LncRNAs are well-known molecules that regulate functional genes in tumor cells, including in GC. Here, we searched for lncRNAs regulating TRIM37 in GC. Based on GEPIA database and results from qRT-PCR assay, we identified ASB16-AS1 as an upregulated lncRNA in GC, suggesting that the potential association of ASB16-AS1 with GC. According to former studies, lncRNAs can be transcriptionally activated in GC cells by various mechanisms. For example, transcription factor SP1 stimulated lncRNA TINCR transcription in GC [30]. LncRNA HOXC-AS3 was transcriptionally activated by H3K4me3 and H3K27ac in GC cells [31]. Therefore, it is possible that ASB16-AS1 could be transcriptionally activated through certain mechanism in GC cells. However, this study focused on the downstream regulatory mechanism ASB16-AS1.
Notably, we first discovered that ASB16-AS1 positively regulated TRIM37 in GC cells. Previously, ASB16-AS1 has been proven as a facilitator for the proliferation and migration of glioma cells and hepatocellular cancer cells [21, 22]. Consistently, we illustrated that ASB16-AS1 induced GC cell proliferation. Besides, we provided novel data that ASB16-AS1 strengthened stemness and cisplatin resistance in GC cells. However, ASB16-AS1 did not stimulate proliferation, stemness and cisplatin resistance in normal GES-1 cells. These data implied the possibility that ASB16-AS1 participated in GC progression rather than initiation.
Later, we first explained how ASB16-AS1 regulated TRIM37 in GC cells. We identified the main distribution of ASB16-AS1 in the cytoplasm of GC cells, and showed that ASB16-AS1 could not regulate the promoter activity of TRIM37, excluding the transcriptional regulation of ASB16-AS1 on TRIM37. Also, we excluded the direct interaction between ASB16-AS1 and TRIM37 mRNA. LncRNAs have been widely demonstrated to be controllers of their downstream targets by serving as a ceRNA in GC [18, 32, 33]. Formerly, ASB16-AS1 functioned as a ceRNA to regulate miR-1874/FZD4 in hepatocellular cancer [21]. Herein, this study first identified miR-3918 and miR-4676-3p as two downstream miRNAs for ASB16-AS1 in GC. A previous study has reported that miR-3918 can cooperate with its sponge circ-BIRC6 to regulate hepatocellular carcinoma progression [34]. As for miR-4676-3p, it has not been studied in any diseases. Therefore, the present study first linked miR-3918 and miR-4676-3p to ASB16-AS1 in GC cells. Moreover, we proved that miR-3918 and miR-4676-3p were downregulated in GC samples. Importantly, the negative correlation of ASB16-AS1 with miR-3918 and miR-4676-3p was analyzed in GC tissues.
Furthermore, TRIM37 has been reported to activate NF-κB pathway and facilitate the aggressiveness of non-small-cell lung cancer [12]. NF-κB pathway is a common oncogenic pathway in various cancers. For instance, Morin suppresses cell growth in ovarian cancer through inhibition of NF-κB signaling [35]. Ubiquitination of IκBα facilitates p65 nuclear translocation, therefore triggering NF-κB pathway activation and inducing EMT process in colorectal cancer cells [36]. MiR-204-5p inactivates NF-κB signaling to repress bone metastasis in prostate cancer [37]. ZBTB20 promotes GC cell migration and invasion through inhibiting IκBα to trigger NF-κB pathway [38]. Currently, we first showed that ASB16-AS1 activated NF-κB pathway in GC cells. Statistics showed that co-inhibition of miR-3918 and miR-4676-3p fully rescued the repressive effect of depleted ASB16-AS1 on TRIM37 level, indicating that miR-3918 and miR-4676-3p might be the main targets for ASB16-AS1 to modulate TRIM37 in GC cells. In the meantime, the partial recovery of NF-κB pathway activity by co-suppressed miR-3918 and miR-4676-3p in GC cells with ASB16-AS1 knockdown suggested that ASB16-AS1 might have alternative ways to regulate NF-κB pathway.
Herein, we stick to the regulation on TRIM37, so we wondered whether ASB16-AS1 regulated TRIM37 through other mechanisms. Our data preliminarily excluded the possibility that ASB16-AS1 regulated TRIM37 mRNA and protein stability. Interestingly, we identified ATM as a protein interacting with ASB16-AS1 in GC cells. ATM has been reported to be an inducer of TRIM37 phosphorylation [26]. Present study first verified that ASB16-AS1 interacted with ATM to induce TRIM37 phosphorylation. Finally, we proved that TRIM37 mediated the influence of ASB16-AS1 on cell proliferation, stemness, drug resistance and tumor growth in GC cells.
In conclusion, the present study first demonstrated that ASB16-AS1 enhanced TRIM37 expression through sequestering miR-3918/miR-4676-3p and facilitated TRIM37 phosphorylation via recruiting ATM. Afterwards, ASB16-AS1 depended on TRIM37 to activate NF-κB pathway to facilitate cell proliferation, stemness, cisplatin resistance and tumor growth in GC. These findings possibly offered new inspirations to GC treatment. However, the upregulation of ASB16-AS1 in GC cells was not clearly explained in this study, which is also a limitation of our current study. We will put more efforts to further explain the regulation of ASB16-AS1 on TRIM37 and other potential targets in GC.
References
Smyth EC, Cunningham D. Gastric cancer in 2012: defining treatment standards and novel insights into disease biology. Nat Rev Clin Oncol. 2013;10(2):73–4. https://doi.org/10.1038/nrclinonc.2012.228(Epub 2013/01/09).
Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–25. https://doi.org/10.1038/bjc.2013.233(Epub 2013/05/11).
Maruyama R, Suzuki H, Yamamoto E, Imai K, Shinomura Y. Emerging links between epigenetic alterations and dysregulation of noncoding RNAs in cancer. Tumour Biol J Int Soc Oncodev Biol Med. 2012;33(2):277–85. https://doi.org/10.1007/s13277-011-0308-9(Epub 2012/01/06).
Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3:17036. https://doi.org/10.1038/nrdp.2017.36(Epub 2017/06/02).
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. https://doi.org/10.3322/caac.21338(Epub 2016/01/26).
Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356(2 Pt B):357–66. https://doi.org/10.1016/j.canlet.2014.11.005(Epub 2014/12/03).
Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–82. https://doi.org/10.1016/j.omtn.2018.09.026(Epub 2018/10/06).
Xu R, Zhu X, Chen F, Huang C, Ai K, Wu H, et al. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018;18:41. https://doi.org/10.1186/s12935-018-0540-0.
An Y, Wang B, Wang X, Dong G, Jia J, Yang Q. SIRT1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an AMPK/FOXO3 positive feedback loop. Cell Death Dis. 2020;11(2):115. https://doi.org/10.1038/s41419-020-2308-4.
Peng X, Liu G, Peng H, Chen A, Zha L, Wang Z. SOX4 contributes to TGF-β-induced epithelial-mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis. 2017;5(1):49–61. https://doi.org/10.1016/j.gendis.2017.12.005.
Tang SL, Gao YL, Wen-Zhong H. Knockdown of TRIM37 suppresses the proliferation, migration and invasion of glioma cells through the inactivation of PI3K/Akt signaling pathway. Biomed Pharmacother Biomedecine Pharmacotherapie. 2018;99:59–64. https://doi.org/10.1016/j.biopha.2018.01.054(Epub 2018/01/13).
Li Y, Deng L, Zhao X, Li B, Ren D, Yu L, et al. Tripartite motif-containing 37 (TRIM37) promotes the aggressiveness of non-small-cell lung cancer cells by activating the NF-kappaB pathway. J Pathol. 2018;246(3):366–78. https://doi.org/10.1002/path.5144(Epub 2018/07/26).
Hu CE, Gan J. TRIM37 promotes epithelialmesenchymal transition in colorectal cancer. Mol Med Rep. 2017;15(3):1057–62. https://doi.org/10.3892/mmr.2017.6125(Epub 2017/01/19).
Tao Y, Xin M, Cheng H, Huang Z, Hu T, Zhang T, et al. TRIM37 promotes tumor cell proliferation and drug resistance in pediatric osteosarcoma. Oncol Lett. 2017;14(6):6365–72. https://doi.org/10.3892/ol.2017.7059(Epub 2017/11/23).
Chen D, You X, Pan Y, Liu Q, Cao G. TRIM37 promotes cell invasion and metastasis by regulating SIP1-mediated epithelial-mesenchymal transition in gastric cancer. OncoTargets Ther. 2018;11:8803–13. https://doi.org/10.2147/ott.s178446(Epub 2018/12/24).
He X, Wang J, Chen J, Han L, Lu X, Miao D, et al. lncRNA UCA1 predicts a poor prognosis and regulates cell proliferation and migration by repressing p21 and SPRY1 expression in GC. Mol Ther Nucleic Acids. 2019;18:605–16. https://doi.org/10.1016/j.omtn.2019.09.024(Epub 2019/11/07).
Liang Y, Zhang CD, Zhang C, Dai DQ. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastr Cancer Off J Int Gastr Cancer Assoc Jpn Gastr Cancer Assoc. 2019. https://doi.org/10.1007/s10120-019-01002-1(Epub 2019/08/30).
Zong W, Feng W, Jiang Y, Cao Y, Ke Y, Shi X, et al. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastr Cancer Off J Int Gastr Cancer Assoc Jpn Gastr Cancer Assoc. 2019. https://doi.org/10.1007/s10120-019-00998-w(Epub 2019/08/28).
Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019;443:67–79. https://doi.org/10.1016/j.canlet.2018.11.032(Epub 2018/12/07).
He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38(23):4637–54. https://doi.org/10.1038/s41388-019-0747-0(Epub 2019/02/11).
Yao X, You G, Zhou C, Zhang D. LncRNA ASB16-AS1 promotes growth and invasion of hepatocellular carcinoma through regulating miR-1827/FZD4 axis and activating Wnt/β-catenin pathway. Cancer Manag Res. 2019;11:9371–8. https://doi.org/10.2147/CMAR.S220434.
Zhang D, Zhou H, Liu J, Mao J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. Biomed Res Int. 2019;2019:5437531. https://doi.org/10.1155/2019/5437531(Epub 2019/04/06).
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. https://doi.org/10.1093/nar/gkt1248(Epub 2013/12/04).
Zheng J, Zhang H, Ma R, Liu H, Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene. 2019. https://doi.org/10.1038/s41388-019-0934-z.
Luo Y, Wu J, Wu Q, Li X, Wu J, Zhang J, et al. miR-577 regulates TGF-β induced cancer progression through a SDPR-modulated positive-feedback loop with ERK-NF-κB in gastric cancer. Mol Ther. 2019;27(6):1166–82. https://doi.org/10.1016/j.ymthe.2019.02.002.
Wu G, Song L, Zhu J, Hu Y, Cao L, Tan Z, et al. An ATM/TRIM37/NEMO axis counteracts genotoxicity by activating nuclear-to-cytoplasmic NF-kappaB signaling. Cancer Res. 2018;78(22):6399–412. https://doi.org/10.1158/0008-5472.can-18-2063(Epub 2018/09/27).
Wang C, Shi M, Ji J, Cai Q, Jiang J, Zhang H, et al. A self-enforcing HOXA11/Stat3 feedback loop promotes stemness properties and peritoneal metastasis in gastric cancer cells. Theranostics. 2019;9(25):7628–47. https://doi.org/10.7150/thno.36277(Epub 2019/11/07).
Lin L, Xiao J, Shi L, Chen W, Ge Y, Jiang M, et al. STRA6 exerts oncogenic role in gastric tumorigenesis by acting as a crucial target of miR-873. J Exp Clin Cancer Res CR. 2019;38(1):452. https://doi.org/10.1186/s13046-019-1450-2(Epub 2019/11/07).
Liu H, Liu N, Zhao Y, Zhu X, Wang C, Liu Q, et al. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging. 2019. https://doi.org/10.18632/aging.102410(Epub 2019/11/07).
Zhang E, He X, Zhang C, Su J, Lu X, Si X, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19(1):154. https://doi.org/10.1186/s13059-018-1523-0.
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–61. https://doi.org/10.1038/onc.2015.18.
Xu TP, Ma P, Wang WY, Shuai Y, Wang YF, Yu T, et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26(11):2179–93. https://doi.org/10.1038/s41418-018-0236-y(Epub 2019/02/17).
Huang J, Chen YX, Zhang B. IGF2-AS affects the prognosis and metastasis of gastric adenocarcinoma via acting as a ceRNA of miR-503 to regulate SHOX2. Gastr Cancer Off J Int Gastr Cancer Assoc Jpn Gastr Cancer Assoc. 2019. https://doi.org/10.1007/s10120-019-00976-2(Epub 2019/06/12).
Yang G, Wang X, Liu B, Lu Z, Xu Z, Xiu P, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle (Georget, Tex). 2019;18(9):976–89. https://doi.org/10.1080/15384101.2019.1601477(Epub 2019/04/02).
Xu M, Zhang Y. Morin inhibits ovarian cancer growth through inhibition of NF-kappaB signaling pathway. Anti Cancer Agents Med Chem. 2019. https://doi.org/10.2174/1871521409666191014164742(Epub 2019/10/30).
Xiao C, Wu G, Zhou Z, Zhang X, Wang Y, Song G, et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial-mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 2019;10(11):833. https://doi.org/10.1038/s41419-019-2070-7(Epub 2019/11/07).
Wa Q, Huang S, Pan J, Tang Y, He S, Fu X, et al. miR-204-5p represses bone metastasis via inactivating NF-kappaB signaling in prostate cancer. Mol Ther Nucleic Acids. 2019;18:567–79. https://doi.org/10.1016/j.omtn.2019.09.008(Epub 2019/11/05).
Zhang Y, Zhou X, Zhang M, Cheng L, Zhang Y, Wang X. ZBTB20 promotes cell migration and invasion of gastric cancer by inhibiting IkappaBalpha to induce NF-kappaB activation. Artif Cells Nanomed Biotechnol. 2019;47(1):3862–72. https://doi.org/10.1080/21691401.2019.1670188(Epub 2019/09/27).
Acknowledgements
We sincerely appreciate all lab members.
Funding
This study was supported by Science Foundation of Peking University Cancer Hospital 2020-12.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethics approval and consent to participate
With the informed consents signed by all patients, this study had received the approval of the Ethics Committee of Peking University Cancer Hospital and Institute.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. A.
qRT-PCR detected the knockdown efficiency of ASB16-AS1, RP11-475B2.1 and AC010518.3. B-C. qRT-PCR and western blot detected TRIM37 expression with RP11-475B2.1 or AC010518.3 silence. D. qRT-PCR of ASB16-AS1 overexpression under pcDNA3.1-ASB16-AS1 transfections. E. qRT-PCR and western blot of TRIM37 level under ASB6-AS1 overexpression in GC cells and normal cells. **P < 0.01, n.s. suggested no significance. (TIFF 2443 kb)
Figure S2 A.
CCK-8 data of GC cell viability under ASB16-AS1 overexpression. B. Pictures of colonies from GC cells with ASB16-AS1 overexpression were taken and colony number was counted. C. Spheres of GC cells were pictured and sphere-forming efficiency was determined. Scale bar: 100 μm. D. CD133+ cell ratio of GC cells with ASB16-AS1 overexpression was measured by flow cytometry analysis. E. CCK-8 data of GC cells transfected with pcDNA3.1 or pcDNA3.1-ASB16-AS1 under the treatment of cisplatin at 2, 4, and 8 μM relative to 0 μM. F. Apoptosis ratio of GC cells with ASB16-AS1 overexpression was detected by flow cytometry under cisplatin treatment. **P < 0.01. (TIFF 1071 kb)
Figure S3 A.
CCK-8 data of normal GES-1 cell viability under ASB16-AS1 overexpression. B. Pictures of colonies from normal GES-1 cell with ASB16-AS1 overexpression were taken and colony number was counted. C. Spheres of normal GES-1 cell were pictured and sphere-forming efficiency was calculated. Scale bar: 100 μm. D. CD133+ cell ratio of normal GES-1 cell with ASB16-AS1 overexpression was measured by flow cytometry analysis. E. CCK-8 data of normal GES-1 cell transfected with pcDNA3.1 or pcDNA3.1-ASB16-AS1 under the treatment of cisplatin at 2, 4, and 8 μM relative to 0 μM. F. Apoptosis ratio of normal GES-1 cell with ASB16-AS1 overexpression was detected by flow cytometry under cisplatin treatment. **P < 0.01, n.s. suggested no significance. (TIFF 831 kb)
Figure S4 A.
Luciferase reporter assay detected the luciferase activity of reporters containing TRIM37 promoter. B. RNA pull-down assay detected the enrichment of TRIM37 mRNA pulled down by ASB16-AS1. C. qRT-PCR examination revealed the expression of 12 candidate miRNAs in GC tissues versus adjacent normal ones. D. qRT-PCR examined miR-3918 and miR-4676-3p expressions in GC cells versus GES-1 cells. E. Correlation between ASB16-AS1 and miR-3918/miR-4676-3p was analyzed based on TCGA data and determined by Pearson correlation analysis in 76 GC samples. F. qRT-PCR verified overexpression efficiency of miR-3918 and miR-4676-3p. *P < 0.05, **P < 0.01, n.s. suggested no significance. (TIFF 738 kb)
Figure S5 A.
qRT-PCR measured TRIM37 mRNA stability after ASB16-AS1 inhibition in GC cells treated with ActD. B. Western blot analysis of TRIM37 protein stability after ASB16-AS1 inhibition in GC cells with CHX treatment. C. p-TRIM37 level in GC cells and GES-1 cells was detected by western blot under ASB16-AS1 overexpression. D. qRT-PCR verified depletion efficiency of ATM. **P < 0.01, n.s. suggested no significance. (TIFF 404 kb)
Rights and permissions
About this article
Cite this article
Fu, T., Ji, K., Jin, L. et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer 24, 45–59 (2021). https://doi.org/10.1007/s10120-020-01096-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-020-01096-y