Abstract
Background
We aimed to identify novel tumor-promoting drivers highly expressed in gastric cancer (GC) that contribute to worsened prognosis in affected patients.
Methods
Genes whose expression was increased and correlated with worse prognosis in GC were screened using datasets from the Cancer Genome Atlas and Gene Expression Omnibus. We examined Claudin-6 (CLDN6) immunoreactivity in GC tissues and the effect of CLDN6 on cellular functions in GC cell lines. The mechanisms underlying GC-promoting function of CLDN6 were also investigated.
Results
CLDN6 was identified as a gene overexpressed in GC tumors as compared with adjacent non-tumorous tissues and whose increased expression was positively correlated with worse overall survival of GC patients, particularly those with Lauren’s intestinal type GC, in data from multiple publicly available datasets. Additionally, membranous CLDN6 immunoreactivity detected in intestinal type GC tumors was correlated with worse overall survival. In CLDN6-expressing GC cells, silencing of CLDN6 inhibited cell proliferation and migration/invasion abilities, possibly via suppressing transcription of YAP1 and its downstream transcriptional targets at least in part.
Conclusions
This study identified CLDN6 as a GC-promoting gene, suggesting that CLDN6 to be a possible single prognostic marker and promising therapeutic target for a subset of GC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed cancer and third leading cause of cancer death worldwide [1]. Despite important advances for clarification of the etiology and molecular basis, as well as development of treatment strategies, survival rates for affected patients remain poor [2]. Presently, two molecular targets, human epidermal growth factor receptor-2 (HER2) and vascular endothelial growth factor receptor-2 (VEGFR2), are available for clinical therapy [3, 4]. However, the heterogeneous nature of GC renders those as only weakly predictive and the subset of patients that seems to benefit from therapies targeting them is small [5]. Therefore, identification of novel prognostic markers and/or therapeutic target genes for better treatment guided by stratification of GC patients is urgently needed to overcome the biological complexity of this disease and maximize outcomes.
Histological classification per se is not enough to explain the high complexity of GC [6]. Recent technical advances along with the efforts of international research consortiums, such as the Cancer Genome Atlas (TCGA) Research Network and Asian Cancer Research Group (ACRG), have led to remarkable progress in elucidation of the genomic landscape of GC [7, 8]. On the other hand, classifications of GC provided by the TCGA and ACRG cannot be currently used for patient stratification or selection, because many of the identified mutations remain functionally unknown and undruggable [9]. Variations in gene expression involved in development and progression of GC may be alternative landmarks for identification of novel tumor-promoting genes to overcome the currently limited number of molecular targets for this disease.
Claudin-6 (CLDN6) is one of the 27 members of the CLDN superfamily, located in the cell membrane and associated with tight junctions of cell adhesion, with expression in normal tissues restricted to the early stages of development [10,11,12]. CLDN6 becomes aberrantly activated in various human cancers including GC [13,14,15,16,17,18], but its clinical and biological relevance is poorly understood.
As an attempt to identify novel tumor-promoting genes involved in GC, we screened differentially overexpressed genes in tumor samples and their prognostic impact using data presented in multiple publicly available datasets from the TCGA and Gene Expression Omnibus (GEO). Those results identified CLDN6 as a gene with one of the greatest amounts of upregulation in GC tumors as compared with non-tumorous tissues as well as an independent prognostic factor for worse overall survival (OS), particularly in patients with Lauren’s intestinal type [19]. In addition, our functional analyses demonstrated growth and/or migration promotion effects of CLDN6 towards GC cells. Together, these findings suggest that CLDN6 is a single prognosticator and functions as an oncogene in at least some GC patient subgroups.
Methods
Data sources and processing
RNA-sequencing (RNA-seq) data normalized by use of the Expectation–Maximization (RSEM) software package and related clinical information for GC patients were obtained from TCGA Research Network (https://cancergenome.nih.gov, discovery cohort). RNA-seq data of paired tumor/non-tumorous tissue samples and those of tumor samples with survival data were available from 31 and 394 GC cases, respectively. To validate the prognostic potential of CLDN6 mRNA expression, four independent datasets (Table S1, validation cohort) containing gene expression profile data from primary GC with patients’ survival data were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Comparisons of differentially expressed genes (DEGs) between matched tumor and non-tumorous tissues were performed using the DESeq2 package (https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) [20]. Adjusted P values were determined using the Benjamini–Hochberg method [21], and values for fold change (FC) > 2 and false discovery rate (FDR) < 0.05 were considered to indicate significance. For analyses of associations between gene expression and OS in the discovery cohort, the 394 cases were divided into two groups based on the median expression level of each gene, then compared using the Kaplan–Meier method with a log-rank test and Cox proportional hazard regression models. In additional analyses of associations between CLDN6 mRNA expression levels and OS in both the TCGA and GEO datasets, a minimum P value approach was employed to find the optimal cutoff point in continuous gene expression measurements for grouping patients. Patients ordered by the expression level of CLDN6 mRNA were divided into two groups at all potential cutoff points and the risk differences of the groups were estimated with a log-rank test. The optimal cutoff point giving the most pronounced P value was selected [22].
Cell lines and primary tissue samples
A total of 15 GC cell lines were used. Twelve lines, including KATOIII, IM95, MKN1, MKN7, MKN45, MKN74, NUGC-2, NUGC-3, NUGC-4, OCUM-1, RERF-GC-1B, and Takigawa, were provided by the Japanese Collection of Research Bioresources (Ibaraki, Japan), while HGC-27 and SH-10-TC were provided by Cell Bank, RIKEN BioResource Center (Tsukuba, Japan), and AGS by the American Type Culture Collection (Manassas, VA, USA).
GC tumor specimens were obtained from 208 patients with histologically proven primary GC staged as pT1-4, pN0-3, M0 who underwent a gastrectomy procedure at the Kyoto Prefectural University of Medicine Hospital between 2009 and 2013 (KPUM cohort). The samples were embedded in paraffin after 24 h of fixation in 10% buffered formalin. None of the patients had synchronous or metachronous multiple cancer in other organs. Relevant clinical and survival data were available for all cases. Disease stage was defined in accordance with the tumor-lymph node-metastasis (TNM) classification of the International Union against Cancer [23]. The median follow-up period for the surviving patients was 57.1 months (range 0.5–60.0 months). Formal written consent was obtained from all patients after receiving approval for all aspects of this study from the ethics committee of Kyoto Prefectural University of Medicine.
Antibodies
Antibodies used in this study are listed in Table S2.
Immunohistochemical staining (IHC) and scoring
Paraffin sections (4-μm thick) were subjected to IHC using DAKO EnVision + Kit/HRP (Agilent Technologies, Santa Clara, CA, USA) for color development with diaminobenzidine tetrahydrochloride, as previously described [24].
Tumor tissues were compared with non-tumorous tissues in each case. The percentage of the total cell population expressing CLDN6 and overall staining intensity in tumor cells were evaluated using images at 200 × magnification. Membranous staining of CLDN6 was considered positive when the cells exhibited some evidence of staining as compared with non-tumorous stomach epithelial cells. CLDN6 expression in tumors was considered positive when over 10% of examined tumor cells showed strong or diffuse staining. All stained slides were evaluated independently in a blinded manner by two different investigators who had no knowledge of the clinicopathological data and any discrepant cases were resolved by consensus review.
Quantitative reverse transcription-PCR (qRT-PCR)
For quantification of mRNA levels, qRT-PCR was performed as previously described using specific primer sets with SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA) or a TaqMan kit (Applied Biosystems) (Table S3) [25]. Human stomach total RNA (Takara Bio, Kusatsu, Japan) was used as a normal stomach tissue. For normalization, the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control.
Western blot analysis
Whole-cell lysate preparations and western blot analysis for each protein (Table S2) were performed with GAPDH used as a loading control, as described in a previous report [24]. Images were obtained with a GE Amersham Imager 600 (GE Healthcare, Milwaukee, WI, USA) or FUSION SOLO.7S.EDGE (Vilber-Lourmat, Marne la Vallée, France).
Fluorescent immunocytochemistry (FIC)
FIC was performed as previously described [24].
Transient transfection experiments
Small interfering RNAs (siRNAs) targeting mRNA of CLDN6 or control siRNA (Table S4) were transfected into cells at a final concentration of 10 nM using Lipofectamine RNAiMax reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
Cell proliferation and cell cycle analysis
Cell proliferation at various times after seeding (1 × 104 cells/24-well plate) was assessed using a water-soluble tetrazolium salt assay (Cell Counting Kit-8; Dojindo Laboratories, Mashikimachi, Japan), according to the manufacturer’s instructions. Results are expressed as the mean absolute absorbance at the indicated time divided by the mean absolute absorbance of each sample cultured for 24 h after seeding.
Cell cycle distribution was determined using fluorescence-activated cell sorting (FACS) with a Muse Cell Analyzer (Merck Millipore, Darmstadt, Germany), according to the manufacturer’s instructions. Obtained data were converted to FCS files using FCS3 Converter 1.0 (Merck Millipore) and analyzed using the Kaluza software package, v.1.5a (Beckman Coulter, Brea, CA, USA).
Transwell migration and invasion assays
Transwell migration and invasion assays were performed using 24-well modified Boyden chambers (Greiner Bio-One GmbH, Frickenhausen, Germany) precoated without or with Matrigel (BD Transduction, Franklin Lakes, NJ, USA), respectively, as previously described [25]. Transfectants (1.0 × 105 cells/well) were transferred into the upper chamber and incubation was performed for 48 h, after which the number of stained cell nuclei on the lower surface of the filter were counted, with the examinations performed in triplicate. The migration and invasive potentials of each transfectant were assessed by calculating the ratio of percentage as compared with the control counterpart.
Expression array analysis
Genome-wide mRNA expression data were obtained from control and CLDN6 knockdown AGS cells using a SuperPrint G3 Human GE 8 × 60 k Microarray (Agilent Technologies), as described elsewhere [26]. All microarray data are available in the GEO database (GSE131787).
Normalized expression data of 42,534 probes were applied to gene set enrichment analysis (GSEA) using the GSEA software package, v.3.0 (https://software.broadinstitute.org/gsea/login.jsp) with oncogenic gene sets from Collection 6 (C6) in the Molecular Signatures Database (MSigDB) 6.2 (https://software.broadinstitute.org/gsea/msigdb) used as the referenced gene sets [27]. Statistical significance of the enrichment score was performed with a permutation test (default = 1000 times). Significance for the gene sets was defined as FDR < 0.1.
Sets of genes showing differential expression with > 2-fold changes in CLDN6-knockdown cells relative to their control counterparts were identified as DEGs. Estimation of potential transcriptional regulators showing binding around the transcription start sites of DEGs was performed using ChIP-Atlas (https://chip-atlas.org/) [28].
Statistical analysis
Clinicopathological variables pertaining to the corresponding patients were analyzed using Fisher’s exact test. For survival analysis, Kaplan–Meier survival curves were constructed for the groups based on univariate predictors and differences between groups were tested using a log-rank test. Univariate and multivariate survival analyses were performed using the likelihood ratio test of the stratified Cox proportional hazards model. Differences between subgroups were evaluated using Student’s t test and assessed with a two-sided test, with P < 0.05 considered to demonstrate significance. All statistical analyses were performed using R version 3.3.3 (R Project for Statistical Computing, Vienna, Austria).
Results
Identification of putative GC-promoting genes using TCGA dataset
To identify putative GC-promoting genes, we screened autosomal genes satisfying both of the following conditions using a TCGA dataset: (1) expression level higher in tumors as compared with adjacent non-tumorous tissues in 31 paired GC samples and (2) increased expression level in tumors associated with worse OS in 394 patients with GC (Fig. S1). Among candidate 83 genes (Table S5), CLDN6 was the second most differentially overexpressed gene in GC tumors as compared with paired non-tumorous tissues (Fig. 1a). Multivariate Cox proportional hazard analysis, which used gender, age, and pathological stage as covariates, demonstrated that CLDN6 but not FEZF1, the top candidate listed in Table S5, was an independent prognosticator for GC tumors. In addition, CLDN6 encodes a cell surface (membrane) protein, which may be useful as a target for molecular targeted strategies in cancer therapy and diagnosis. Therefore, we focused on CLDN6 in further analyses to elucidate its clinicopathological and functional significance in relation to GC development.
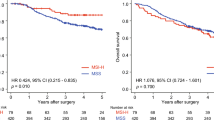
aCLDN6 mRNA expression in 31 GC tumors and paired non-tumorous tissues from TCGA dataset. The y-axis represents the log ratio of RSEM determined by RNA-seq. b Histogram of CLDN6 mRNA expression values for GC patients from TCGA dataset. The cutoff point to discriminate patients with CLDN6-high from those with CLDN6-low GC tumors was determined using the median value of 394 GC samples [log2 (RSEM + 1) = 1.58, median value model] or optimal value that resulted in the most pronounced P value for risk difference between the two groups with a log-rank test [log2 (RSEM + 1) = 5.36, minimum P value model]. c Kaplan–Meier curves for OS rates of 394 GC patients classified into CLDN6-high and -low expression groups according to median value model (described in Fig. 1b). A log-rank test was used for statistical analysis. d Kaplan–Meier curves for OS rates of 394 GC patients classified into CLDN6-high and -low expression groups according to minimum P value model (described in Fig. 1b). e Kaplan–Meier curves for OS rates of 394 GC patients classified into intestinal type with CLDN6-high, intestinal type with CLDN6-low, diffuse type with CLDN6-high, and diffuse type with CLDN6-low expression groups. The cutoff point to discriminate patients with CLDN6-high from those with CLDN6-low GC tumors was determined using the minimum P value model (described in Fig. 1b)
Clinicopathological significance of CLDN6 expression in GC using TCGA dataset
A precise review of the CLDN6 mRNA expression status demonstrated that most GC tumors showed a low CLDN6 expression level, though some showed a remarkably higher expression level (Fig. 1b), suggesting the existence of a small subset of GC cases with highly elevated CLDN6 expression, which was previously shown by IHC findings of GC [13, 29]. Therefore, instead of using median CLDN6 mRNA level, the optimal cutoff point was defined as the point with the most significant split for correlation with OS and used as the cutoff value to divide all samples into two groups for further survival analysis (Fig. 1b, c). Using the optimal cutoff point [log2 (RSEM + 1) = 5.36], all patients were divided into CLDN6-low (n = 323) and -high (n = 71) groups, which resulted in the greatest significant difference of OS (Fig. 1d). Associations between clinicopathological features and CLDN6 mRNA expression status in the TCGA dataset using the optimal cutoff point for division are summarized in Table 1. Notably, most cases in the CLDN6-high group were intestinal type in the Lauren classification, showed the microsatellite stable (MSS) or microsatellite instability-low (MSI-L) phenotype, and were classified as chromosomal instability (CIN) molecular subtype using the TCGA classification [7]. Multivariate Cox-proportional hazard regression analysis identified higher CLDN6 mRNA expression, older age (> 65 years), and Lauren classification (diffuse type) as independent predictive factors for worse OS (Table 2). Similar findings were obtained even in cases with intestinal type GC (Tables S6, S7). By integrating Lauren classification status with CLDN6 mRNA expression status, we then conducted survival analysis among 4 groups (intestinal or diffuse, CLDN6-high or -low) (Fig. 2e). As reported previously [30], cases classified as the intestinal type showed better OS as compared with the diffuse type using Kaplan–Meier survival estimates (Fig. S2a). Among cases with intestinal type GC, the CLDN6-high subgroup had a worse OS rate than the CLDN6-low subgroup, while the CLDN6-high and -low diffuse type subgroups showed similar rates for OS.
a Representative images of immunohistochemically detected CLDN6 protein in normal gastric mucosa and advanced intestinal type GC. In one case with positive CLDN6 immunoreactivity, images with different magnifications were shown. HE stained image with low magnification was also shown in the same case. Scale bars in each image represent indicated length. b Kaplan–Meier curves for OS rates of 208 patients (103 intestinal type and 105 diffuse type GC) classified into CLDN6-positive and -negative expression groups according to CLDN6 immunoreactivity of tumor. c Kaplan–Meier curves for OS rates of 208 GC patients classified into intestinal type with CLDN6-positive, intestinal type with CLDN6-negative, diffuse type with CLDN6-positive, and diffuse type with CLDN6-negative expression groups
Validation of findings in TCGA dataset using GEO datasets
We then validate the findings obtained with the TCGA dataset through pooled analysis using independent microarray data of four cohorts from the GEO datasets (Table S1). Similar to the findings in the TCGA dataset, only a part of the GC tumors showed a high level of CLDN6 mRNA expression (Fig. S3a). Using the optimal cutoff point with the most significant split for correlation with OS, we obtained results similar to those from the TCGA dataset (Table S8). More intestinal type cases showed a higher level of CLDN6 mRNA expression as compared with diffuse or mixed type cases, though no statistically significant difference was observed among the subgroups. In all cases or those with intestinal type GC, the CLDN6-high subgroup showed worse OS than the CLDN6-low subgroup (Fig. S3b). Furthermore, though intestinal type cases showed better OS as compared with diffuse type (Fig. S2b), the CLDN6-high subgroup with intestinal type showed the worst OS (Fig. S3c). Multivariate Cox-proportional hazard regression analysis identified higher CLDN6 mRNA expression and pathologic stage (stage II–IV) as independent predictive factors for worse OS (Table S9).
Immunohistochemical analysis of CLDN6 expression in GC
Next, we performed IHC using a CLDN6-specific antibody with 208 surgically resected GC samples (Fig. 2a). CLDN6 immunoreactivity was not observed in non-tumorous epithelia from any of those cases or in cancer cells from 180 of the GC samples. However, in 28 samples, membranous CLDN6 immunoreactivity was heterogeneously observed in tumor cells, with that immunoreactivity sometimes greater in tumor cells located in the invasive front as compared with those in the center of the tumor.
CLDN6 immunoreactivity was significantly associated with pN category and pathologic stage in the TNM classification and Lauren classification (Table S10). Kaplan–Meier survival estimates showed that positive CLDN6 immunoreactivity in tumor cells was significantly associated with a worse OS in all GC cases (Fig. 2b). Among cases with intestinal type GC, the CLDN6-positive subgroup had a worse OS rate than the CLDN6-negative subgroup, while the CLDN6-positive and -negative diffuse type subgroups showed similar rates for OS (Fig. 2c), although cases with intestinal and diffuse type GC showed similar rate for OS (Fig. S2c). Using a Cox proportional hazard regression model, univariate analyses demonstrated that CLDN6 immunoreactivity, age, and pathologic stage of TNM classification were significantly associated with OS (Table S11). When the data were stratified for multivariate analysis using Cox regression procedures, only age and pathologic stage remained significant for OS. Similar findings were obtained even in cases with intestinal type GC (Tables S12, S13).
Knockdown of CLDN6 suppresses GC cell proliferation, migration, and invasion
Relatively higher CLDN6 mRNA expression was detected in three cell lines, AGS, MKN7, and NUGC-3, of the 15 GC cell lines, but not detected in normal stomach tissues by qRT-PCR (Fig. 3a). AGS and NUGC-3 cell lines were derived from poorly differentiated carcinomas, whereas MKN7 cell line was derived from differentiated carcinomas showing morphological characteristics of intestinal differentiation. With FIC staining, a larger fraction of endogenously expressed CLDN6 protein was found in the plasma membrane, especially in areas of cell–cell contact, in those cell lines (Fig. 3b). Therefore, we used those for further analyses to gain insight into the potential function of CLDN6, as its overexpression was considered to be possibly associated with the malignant phenotype of GC.
a Expression levels of CLDN6 mRNA in a panel of GC cell lines and the normal stomach tissue (NT) were determined using qRT-PCR and normalized by GAPDH. Values are expressed as fold change (mean ± SD, N = 4) as compared with values for the Takigawa cell line (mean ± SD, N = 4). n.d. not detected. b Representative images of AGS, MKN7, and NUGC-3 cells subjected to FIC using an anti-CLDN6 antibody as a primary antibody (green). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Scale bar 50 μm. c Protein expression levels of p21WAF1/Cip1, p27Kip1, SKP2, CDH1, and SNAI1 in GC cell lines with a relatively high level of expression of endogenous CLDN6 after treatment with 10 nM control (−) or CLDN6-specific siRNAs
First, we examined the effects of CLDN6 knockdown on cell proliferation. By treating with two different siRNAs (Fig. 3c), cell proliferation was significantly suppressed in AGS, MKN7, and NUGC-3 cells (Fig. 4a). Using FACS analysis, an accumulation of cells in the G0–G1 phase and a decrease in those in the S and G2–M phases was observed among CLDN6 siRNA-treated cells as compared with control siRNA-treated cells (Fig. 4b). Knockdown of endogenous CLDN6 significantly increased p21WAF1/Cip1 and p27Kip1, and decreased SKP2 protein levels, each of which is a well-known cell cycle regulator (Fig. 3c). These results indicated that CLDN6 silencing in GC cells contributes to cell cycle arrest at the G1–S checkpoint.
a GC cells were transfected with 10 nM of the control or CLDN6-specific siRNAs for 24 h, then cell proliferation was determined using a WST-8 assay at the indicated times. Values are expressed as fold changes (mean ± SD, N = 6) as compared with the respective values for the control cells (0 h). *P < 0.05. b Representative results of cell cycle analysis by FACS using GC cells after treatment with 10 nM CLDN6-specific or control siRNA for 48 h. Raw data were quantified using the Kaluza software package (v.1.5a). c GC cells were treated as described in Fig. 4a, then placed in Boyden chambers precoated without (migration assay, left) or with Matrigel (invasion assay, right). Following incubation for 48 h, the number of cells on the lower surface of the filter was counted as described in the “Materials and methods” section (mean ± SD, N = 6). *P < 0.05
We next assessed the effects of CLDN6 knockdown on cell migration and invasion abilities using Transwell assays. In three cell lines, the number of CLDN6 siRNA-transfected cells that migrated into the lower chamber through an uncoated membrane was significantly lower as compared with the control cells (Fig. 4c). Since MKN7 and NUGC-3 cells showed a low amount of invasion, we used AGS cells for invasion assays. The difference in invasion ability of those three cell lines might be explained by different expression levels of endogenous CDH1 and SNAI1 (Fig. 3c), which are negative and positive markers, respectively, of epithelial-to-mesenchymal transition (EMT). In the AGS cell line, the number of cells that moved to the lower chamber through a Matrigel-coated membrane was reduced by CLDN6 knockdown. In western blot analysis, CLDN6 knockdown induced an increase in CDH1 protein expression in NUGC-3 and a decrease in SNAI1 protein expression in AGS and MKN7 cells (Fig. 3c), suggesting that EMT may also be inhibited by CLDN6 knockdown.
CLDN6 knockdown suppresses transcription of YAP1 and its transcriptional targets
To better elucidate the molecular mechanisms of the tumor-promoting function of CLDN6, we performed expression-array analysis to determine the effects of CLDN6 knockdown on the AGS cell transcriptome.
We initially applied GSEA to detect the signatures of oncogenic pathway activation gene sets (C6) correlated with CLDN6 expression status, and identified 19 sets significantly enriched in control cells as compared with the CLDN6-knockdown cells (Table S14). The YAP1 conserved signature named ‘CORDENONSI YAP CONSERVED SIGNATURE’ was the most significantly enriched (Fig. 5a), though other signatures related with cell proliferation, migration, and invasion, e.g., signatures of genes positively regulated by E2F1, MEK, and mTOR, and negatively by RB, were also enriched. We then screened the functional downstream modules related to 804 differentially downregulated genes based on the criterium of at least a twofold change in CLDN6-knockdown cells as compared with the control cells by estimating enriched potential transcriptional regulators, which bind around transcription start sites of these differentially downregulated genes, using the ChIP-Atlas. Among transcription factors or cofactors whose targets were significantly downregulated by CLDN6 silencing, components of the Hippo signaling pathway transducer YAP/TAZ-TEAD complex, YAP1, TEAD1, and TEAD4 [31,32,33], were found (Table S15). Because results of two different analyses demonstrated that the downstream molecules of the YAP/TAZ-TEAD complex were enriched as downregulated genes by CLDN6 knockdown at the transcript level and the expression-array analysis detected YAP1 as the only molecule whose mRNA level was significantly downregulated by CLDN6 knockdown among components of the YAP/TAZ-TEAD complex, we further focused on YAP1 and transcriptional targets of the YAP/TAZ-TEAD complex. CLDN6 knockdown-induced decreases in YAP1 mRNA and protein were validated by qRT-PCR and western blot analysis results, respectively (Fig. 5b, c). In addition, CLDN6 knockdown-induced decreases of several known cancer-related genes transcriptionally regulated by the YAP/TAZ-TEAD complex, such as ANKRD1, CTGF, CYR61, and EDN1 [31,32,33], were detected at mRNA level, although smaller or the opposite effects of CLDN6 knockdown were observed in MKN7 cells compared with other two cell lines (Fig. 5a, c).
Effects of CLDN6 knockdown on YAP1 and its downstream target molecules in GC cell lines. a Heat map of gene expression changes in the gene set 'CORDENONSI YAP CONSERVED SIGNATURE' in GSEA analysis when comparing CLDN6-knockdown AGS cells with the control cells. Colors range from dark red to dark blue representing respectively the highest and lowest expression of a gene. Arrowheads indicate genes whose expression was validated by qRT-PCR in AGS, MKN7, and NUGC-3 cells as shown in Fig. 5c. b YAP1 protein levels in CLDN6 knockdown cells were determined by western blot analysis. c Expression levels of ANKRD1, CYR61, CTGF, EDN1, and YAP1 mRNAs were determined by qRT-PCR and normalized by GAPDH. Values are expressed as fold change (mean ± SD, N = 4) as compared with the respective values obtained with control siRNA-transfected cells. *P < 0.05. d Model depicts the possible mechanism whereby CLDN6 promotes proliferation and migration/invasion of GC cells, though signaling pathways from CLDN6 to transcriptional regulators for YAP1 remain unknown
Discussion
In the present study, we demonstrated that high CLDN6 expression observed in a subset of GC tumors, particularly those from intestinal type GC cases, is associated with worse OS. Recently, CLDN6 was reported to be expressed in a subset of GC cases that predominantly consist of intestinal type adenocarcinoma with a fetal gut-like phenotype, as well as to be one of markers for the primitive enterocyte phenotype of GC associated with tumor aggressiveness [29]. Our results suggest the significance of higher CLDN6 expression alone as a biomarker for aggressiveness and/or poor patient prognosis of intestinal type GC.
Results contrary to findings of the present study have been reported for GC as well as several other types of cancers. In GC (1) lower CLDN6 protein [34] and mRNA [35] levels in tumors as compared with non-tumorous tissues, (2) an association of lower CLDN6 mRNA expression in tumors with worse prognosis [35], and (3) an increased CLDN6 protein expression in both intestinal and diffuse types [36] have been reported. In breast cancer patients, tumor-specific downregulation of CLDN6 expression and its association with lymphatic metastasis have also been noted [37]. Although it is possible that CLDN6 has a tissue- or lineage-dependent function in relation to carcinogenesis, substantive reasons for the inconsistent findings obtained in the same cancer type remain unclear. Because most GC tumors and adjacent non-tumorous tissues showed a very low CLDN6 expression level, an erroneous determination/grouping based on heterogeneous CLDN6 expression in tissue samples. In addition, because only a small subset of GC cases shows highly elevated CLDN6 expression (Fig. 1b) and this subset are more frequently observed in intestinal type GC as compared with diffuse type GC (Table 1), the sample sizes of intestinal and diffuse type GC cases may affect the results of analyses. The differences between the present results of mRNA analyses of data from the TCGA/GEO datasets and those of IHC analysis of the KPUM cohort, e.g., independent significance of CLDN6 as a prognostic marker, might be explained in the same way, indicating that further analyses using larger cohorts are needed to determine better analytical methods, as well as cutoff values and definitions for CLDN6 expression status.
CLDN6 is known to be a tight junction membrane protein. Although several of the 27 Claudin molecules including CLDN6 harbor a putative nuclear localization sequence [38], the present IHC and FIC results demonstrated that the endogenous CLDN6 protein is mainly localized in the membrane of GC cells in both primary tumors and cultured cells. In addition, CLDN6 is not expressed in most of normal adult tissues, but expressed in various types of embryonic epithelia [10,11,12]. Therefore, CLDN6 seems to be an ideal target for an antibody-based approach for GC therapy with high potency. Several reagents have been developed and are currently being subjected to evaluation, including a currently ongoing phase I/II trial of IMAB027, an immune effector mobilizing antibody shown to kill tumor cells through antibody-dependent cell-mediated cytotoxicity, for patients with recurrent advanced ovarian cancer [39]. Additionally, highly efficient therapeutic effects of 6PHU3, a T-cell engaging bispecific single-chain molecule with anti-CD3/anti-CLDN6 specificities, on CLDN6-expressing ovarian cancer cells have been reported from results of a preclinical validation [40]. The present investigation revealed the CLDN6 knockdown-induced anti-cancer effects on CLDN6-expressing GC cells, thus reagents that silence expression or inactivate the biological effects of CLDN6 even without mobilization of immune effectors may be effective for CLDN6-expressing aggressive GC tumors. Further developments and clinical trials of novel reagents targeting such tumors are eagerly anticipated.
This study demonstrated accelerated effects of endogenously overexpressed CLDN6 on GC cell proliferation, migration, and invasion. In a previous study using AGS cells, similar effects of CLDN6 towards cell proliferation, migration, and invasion were shown as a result of its exogenous overexpression [41]. AGS is a cell line with relative overexpression of endogenous CLDN6, thus our results with the present CLDN6 knockdown model suggest that endogenously overexpressed CLDN6 may have an essential function as a driver for malignant phenotypes of this cell line. NUGC-3 and MKN7 cell lines, which also show relative overexpression of CLDN6, have a less invasive phenotype possibly due to low expression of endogenous effector molecules essential for an invasive phenotype including SNAI1. In addition, weaker or the opposite effects of CLDN6 knockdown on transcription of YAP1 and its target genes were observed in MKN7 compared with other two cell lines, although a similar effect was observed in the YAP1 protein among three cell lines. These results suggest that the different status of dependency on CLDN6 among cell lines may be determined by endogenous activities of the effector molecules and/or responsiveness of the target molecules required for each phenotype. Indeed, endogenous CLDN6-induced cell proliferation and migration have been reported regarding HEC-1-B, an endometrial carcinoma cell line [42], whereas inhibition of cell migration and invasion by restoration of CLDN6 was shown in the breast cancer cell line MCF-7 [43]. Additional studies are needed to clarify the detailed molecular mechanisms underlying the tumor-promoting activity of CLDN6 in association with GC.
Our expression-array analysis using the CLDN6-knockdown GC cells revealed that CLDN6 may exert tumor-promoting function via activation of the YAP/TAZ-TEAD complex by an increase in YAP1 transcription, at least in part, though the pathways between CLDN6 expressed in the cell membrane and regulators for YAP1 transcription remain unknown (Fig. 5d). In the TCGA and GEO datasets, a small subset of GC tumors with very high CLDN6 mRNA expression tended to show higher YAP1 mRNA expression, although many tumors showed high YAP1 mRNA expression regardless of CLDN6 mRNA expression level (Fig. S4), suggesting that CLDN6 may not always be necessary but one of multiple factors/mechanisms to activate YAP1 transcription. In GC, YAP1 mRNA and protein overexpression, nuclear localization of YAP1, and their prognostic values have been reported previously [44,45,46,47]. Various molecules including microRNAs also have been reported as regulators of YAP1 expression level [48,49,50]. Embryonic-like stemness of cancers, e.g. polyploid giant cancer cells, expressing various embryonic stem cell markers has been reported to be associated with nuclear accumulation of YAP1 [51], suggesting that it will be needed to clarify functional role of YAP1 in GC with primitive enterocytic phenotype.
In conclusion, our systematic and integrative analyses demonstrated that tumor-specific upregulation of CLDN6 expression results in a relatively malignant phenotype, which is mediated, at least in part, through activating YAP1 transcription in GC, particularly a subset of intestinal type cases. Therefore, CLDN6 might be a novel single prognostic marker and promising therapeutic target for a subset of GC patients.
Data availability
All data generated or analyzed during this study are included either in this article or the additional files. All microarray data are available in the GEO database (GSE131787).
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68:394–424.
Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64.
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97.
Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–9.
Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3:17036.
Carrasco-Garcia E, Garcia-Puga M, Arevalo S, Matheu A. Towards precision medicine: linking genetic and cellular heterogeneity in gastric cancer. Ther Adv Med Oncol. 2018;10:1758835918794628.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56.
Tirino G, Pompella L, Petrillo A, Laterza MM, Pappalardo A, Caterino M, et al. What's new in gastric cancer: the therapeutic implications of molecular classifications and future perspectives. Int J Mol Sci. 2018;19:2659.
Abuazza G, Becker A, Williams SS, Chakravarty S, Truong HT, Lin F, et al. Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol. 2006;291:F1132–F1141141.
D'Souza T, Sherman-Baust CA, Poosala S, Mullin JM, Morin PJ. Age-related changes of claudin expression in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med Sci. 2009;64:1146–53.
Turksen K, Troy TC. Permeability barrier dysfunction in transgenic mice overexpressing claudin 6. Development. 2002;129:1775–844.
Ushiku T, Shinozaki-Ushiku A, Maeda D, Morita S, Fukayama M. Distinct expression pattern of claudin-6, a primitive phenotypic tight junction molecule, in germ cell tumours and visceral carcinomas. Histopathology. 2012;61:1043–56.
Micke P, Mattsson JS, Edlund K, Lohr M, Jirström K, Berglund A, et al. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int J Cancer. 2014;135:2206–14.
Ben-David U, Nudel N, Benvenisty N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat Commun. 2013;4:1992.
Cherradi S, Martineau P, Gongora C, Del Rio M. Claudin gene expression profiles and clinical value in colorectal tumors classified according to their molecular subtype. Cancer Manag Res. 2019;11:1337–488.
Vare P, Soini Y. Twist is inversely associated with Claudins in germ cell tumors of the testis. APMIS. 2010;118:640–7.
Birks DK, Kleinschmidt-DeMasters BK, Donson AM, Barton VN, McNatt SA, Foreman NK, et al. Claudin 6 is a positive marker for atypical teratoid/rhabdoid tumors. Brain Pathol. 2010;20:140–50.
Lauren P. The Two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Tan YD, Xu H. A general method for accurate estimation of false discovery rates in identification of differentially expressed genes. Bioinformatics. 2014;30:2018–25.
Abel U, Berger J, Wiebelt H. CRITLEVEL: an exploratory procedure for the evaluation of quantitative prognostic factors. Methods Inf Med. 1984;23:154–6.
Sobin LH, Gospodarowicz MK, Wittekind C, International Union against Cancer. TNM classification of malignant tumours. 7th ed. Chichester: Wiley-Blackwell; 2010.
Hamada J, Shoda K, Masuda K, Fujita Y, Naruto T, Kohmoto T, et al. Tumor-promoting function and prognostic significance of the RNA-binding protein T-cell intracellular antigen-1 in esophageal squamous cell carcinoma. Oncotarget. 2016;7:17111–28.
Fujita Y, Masuda K, Hamada J, Shoda K, Naruto T, Hamada S, et al. KH-type splicing regulatory protein is involved in esophageal squamous cell carcinoma progression. Oncotarget. 2017;8:101130–45.
Kajiura K, Masuda K, Naruto T, Kohmoto T, Watabnabe M, Tsuboi M, et al. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget. 2017;8:2890–905.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018;19:e46255.
Yamazawa S, Ushiku T, Shinozaki-Ushiku A, Hayashi A, Iwasaki A, Abe H, et al. Gastric cancer with primitive enterocyte phenotype: an aggressive subgroup of intestinal-type adenocarcinoma. Am J Surg Pathol. 2017;41:989–97.
Petrelli F, Berenato R, Turati L, Mennitto A, Steccanella F, Caporale M, et al. Prognostic value of diffuse versus intestinal histotype in patients with gastric cancer: a systematic review and meta-analysis. J Gastrointest Oncol. 2017;8:148–63.
Ehmer U, Sage J. Control of proliferation and cancer growth by the Hippo signaling pathway. Mol Cancer Res. 2016;14:127–40.
Park JH, Shin JE, Park HW. The role of Hippo pathway in cancer stem cell biology. Mol Cells. 2018;41:83–92.
Liu H, Du S, Lei T, Wang H, He X, Tong R, et al. Multifaceted regulation and functions of YAP/TAZ in tumors (Review). Oncol Rep. 2018;40:16–28.
Lin Z, Zhang X, Liu Z, Liu Q, Wang L, Lu Y, et al. The distinct expression patterns of claudin-2, -6, and -11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol. 2013;8:133.
Gao F, Li M, Xiang R, Zhou X, Zhu L, Zhai Y. Expression of CLDN6 in tissues of gastric cancer patients: association with clinical pathology and prognosis. Oncol Lett. 2019;17:4621–5.
Rendon-Huerta E, Teresa F, Teresa GM, Xochitl GS, Georgina AF, Veronica ZZ, et al. Distribution and expression pattern of claudins 6, 7, and 9 in diffuse- and intestinal-type gastric adenocarcinomas. J Gastrointest Cancer. 2010;41:52–9.
Xu X, Jin H, Liu Y, Liu L, Wu Q, Guo Y, et al. The expression patterns and correlations of claudin-6, methy-CpG binding protein 2, DNA methyltransferase 1, histone deacetylase 1, acetyl-histone H3 and acetyl-histone H4 and their clinicopathological significance in breast invasive ductal carcinomas. Diagn Pathol. 2012;7:33.
Hagen SJ. Non-canonical functions of claudin proteins: beyond the regulation of cell-cell adhesions. Tissue Barriers. 2017;5:e1327839.
Sahin U, Jaeger D, Marme F, Mavratzas A, Krauss J, De Greve J, et al. First-in-human phase I/II dose-escalation study of IMAB027 in patients with recurrent advanced ovarian cancer (OVAR): preliminary data of phase I part. J Clin Oncol. 2015;33(suppl):5537.
Stadler CR, Bahr-Mahmud H, Plum LM, Schmoldt K, Kolsch AC, Tureci O, et al. Characterization of the first-in-class T-cell-engaging bispecific single-chain antibody for targeted immunotherapy of solid tumors expressing the oncofetal protein claudin 6. Oncoimmunology. 2016;5:e1091555.
Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montano LF, Rendon-Huerta EP. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest. 2011;29:1–11.
Cao X, He GZ. Knockdown of CLDN6 inhibits cell proliferation and migration via PI3K/AKT/mTOR signaling pathway in endometrial carcinoma cell line HEC-1-B. Onco Targets Ther. 2018;11:6351–60.
Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res. 2016;35:120.
Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–9.
Song M, Cheong JH, Kim H, Noh SH, Kim H. Nuclear expression of Yes-associated protein 1 correlates with poor prognosis in intestinal type gastric cancer. Anticancer Res. 2012;32:3827–34.
Hu X, Xin Y, Xiao Y, Zhao J. Overexpression of YAP1 is correlated with progression, metastasis and poor prognosis in patients with gastric carcinoma. Pathol Oncol Res. 2014;20:805–11.
Yu L, Gao C, Feng B, Wang L, Tian X, Wang H, et al. Distinct prognostic values of YAP1 in gastric cancer. Tumour Biol. 2017;39:1010428317695926.
Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14:52.
Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:92.
Zhou H, Li G, Huang S, Feng Y, Zhou A. SOX9 promotes epithelial-mesenchymal transition via the Hippo-YAP signaling pathway in gastric carcinoma cells. Oncol Lett. 2019;18:599–608.
Niu N, Mercado-Uribe I, Liu J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 2017;36:4887–900.
Acknowledgements
This study was supported in part by JSPS KAKENHI, grant number JP18H02894 as a Grant-in-Aid for Scientific Research (B) (to I.I.), 18K07910 as a Grant-in-Aid for Scientific Research (C) (to K.M), and 18J21308 as a Grant-in-Aid for JSPS Research Fellow (to T.K.). We thank Hideaki Horikawa and Akiko Watanabe (Support Center for Advanced Medical Sciences, Graduate School of Biomedical Sciences, Tokushima University, Tokushima, Japan) for their technical assistances.
Author information
Authors and Affiliations
Contributions
All of the listed authors contributed to the current study. II conceived and designed the experiments; TK and KM performed the experiments; TK, KM, KS, ST, and II analyzed the data; KS, DI, and EO performed collection of the tissue specimens; and TK, KM, and II wrote the paper. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have conflicts of interest to declare.
Ethics approval
All procedures were performed in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national), as well as the Helsinki Declaration of 1964 and later versions, and approved by the ethics committee of Kyoto Prefectural University of Medicine.
Informed consent
Informed consent or an acceptable substitute was obtained from all patients prior to inclusion in the study.
Consent for publication
Consent to publish the present findings was obtained from all of the participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1.
Outline of strategy used to identify candidate GC-promoting genes with systematic bioinformatic analysis.. Fig. S2. (a) Kaplan–Meier curves for OS rates of 231 GC patients whose Lauren classification data were available from TCGA database. The patients were classified into the intestinal and diffuse groups according to the Lauren criteria. A log-rank test was used for statistical analysis. (b) Kaplan–Meier curves for OS rates of 573 GC patients with Lauren classification data available from the GEO datasets. The patients were classified into the intestinal and diffuse groups according to the Lauren criteria. A log-rank test was used for statistical analysis. (c) Kaplan–Meier curves for OS rates of 208 GC patients from the KPUM cohort. The patients were classified into the intestinal and diffuse groups according to the Lauren criteria. A log-rank test was used for statistical analysis. Fig. S3. (a) Histogram of CLDN6 mRNA expression for GC patients from GEO datasets. The cutoff point to discriminate patients with CLDN6-high from those with CLDN6-low GC tumors was determined using a minimum P value model obtained from log-rank test results of 633 GC samples (normalized signal intensity = 5.10). (b) Kaplan–Meier curves for OS rates of 633 GC patients classified into CLDN6-high and -low expression groups according to values using the method described in Fig. S3a. (c) Kaplan–Meier curves for OS rates of 633 GC patients classified into intestinal type with CLDN6-high, intestinal type with CLDN6-low, diffuse type with CLDN6-high, and diffuse type with CLDN6-low groups. The cutoff value for discriminating patients with CLDN6-high from those with CLDN6-low GC tumors was determined using the method described in Fig. S3a. Fig. S4. Relationship between CLDN6 and YAP1 mRNA expression for GC patients from TCGA (a) and GEO (b) data sets. The horizontal and vertical red dotted lines indicate median of YAP1 and CLDN6 mRNA expression, respectively. Modified histograms of CLDN6 mRNA expression for GC patients from TCGA (Fig. 1b) and GEO (Fig. S3a) data sets are shown below the scatter plots. (PDF 118 kb)
Rights and permissions
About this article
Cite this article
Kohmoto, T., Masuda, K., Shoda, K. et al. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer 23, 403–417 (2020). https://doi.org/10.1007/s10120-019-01014-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-01014-x