Abstract
Background
Few reliable prognostic markers have been established despite elucidation of the molecular mechanisms of gastrointestinal stromal tumor (GIST) development. We evaluated F-box and WD repeat domain-containing 7 (FBXW7), a cell-cycle-regulating and tumor suppressor, in GISTs. We aimed to determine the clinical relevance of FBXW7 in GISTs and characterize the molecular mechanism of FBXW7 in a GIST cell line.
Methods
We measured FBXW7 expression in 182 GIST cases, correlated the expression levels with clinicopathological features, and characterized the molecular mechanism underlying suppressed FBXW7 expression in GIST cells in vitro.
Results
Of the 182 GISTs, 98 (53.8%) and 84 (46.2%) were categorized in the high and low FBXW7 expression groups, respectively. Compared with the high FBXW7 expression group, the low expression group showed a significantly poorer prognosis in terms of recurrence-free (P = 0.01) and overall (P = 0.03) survival. FBXW7 expression was a significant independent factor affecting the 10-year recurrence-free survival rate (P = 0.04). In vitro, FBXW7-specific siRNAs enhanced c-myc and Notch 1 protein expression and upregulated cell proliferation, invasion, and migration.
Conclusion
FBXW7 is a potential predictive marker of recurrence after curative resection of GISTs. FBXW7 expression may help identify patients benefitting from adjuvant therapy more precisely compared with a conventional risk stratification model.
Similar content being viewed by others
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumor of the gastrointestinal tract with an annual incidence of approximately 10–15 cases per million population [1]. GISTs originate from the interstitial cells of Cajal lineage after the development of activating mutations in c-KIT or platelet-derived growth factor receptor alpha [2, 3]. Surgical resection with negative margins is the mainstay of treatment for permanent cure of primary GISTs [4, 5]. However, approximately 50% of patients receiving complete resection subsequently experience recurrence within 5 years after surgery [6]. The success of imatinib, which selectively inhibits the tyrosine kinase c-KIT, for treatment of recurrent or metastatic disease makes it possible for high-risk patients to adapt to adjuvant therapy after curative surgery [7].
The factors responsible for the malignant and aggressive clinical behavior of GISTs have been investigated for years [8]. Several risk classifications for recurrence have been established, including tumor size, mitotic index, tumor location, and tumor rupture [9]. Recently, a contour map was developed to predict GIST recurrence after surgery to help guide patient selection for adjuvant therapy [10]. Although clinical indices such as mitotic count and primary tumor location and size have been used to predict the risk of recurrence, few reliable prognostic molecular biomarkers have been established for GISTs. A recent study demonstrated that expression of cell-cycle regulators such as cyclin D1, CDK4, p21, and p16 is associated with a high risk of GIST recurrence [11]. This finding strongly supports that cell-cycle regulators are associated with the clinicopathological characteristics (e.g., mitotic count and tumor size) that can lead to malignant behavior in GISTs.
Ubiquitin-mediated proteolysis regulates the degradation of many proteins involved in the control of cell differentiation and growth [12]. F-box and WD repeat domain-containing 7 (FBXW7) is the substrate recognition component of an evolutionarily conserved SCF (SKP1–CUL1–F-box protein)-type ubiquitin ligase complex, which is a well-known tumor suppressor that regulates the stability of multiple oncoproteins, including cyclin E, c-myc, and Notch 1 [13]. Regulation of the cell cycle is a critical function of FBXW7, which is associated with tumor progression and cell division [14, 15]. We have demonstrated that low expression of FBXW7 is associated with the poorer prognosis of gastrointestinal cancer via regulation of genes related to cell cycle [16,17,18].
In this study, we hypothesized that FBXW7 plays a key role in tumor progression of GISTs via regulation of tumor size and tumor cell mitosis. To confirm the clinicopathological significance of FBXW7 in GISTs, we examined the expression of FBXW7 in a series of 182 GIST patients with known clinicopathological characteristics, including prognosis. Furthermore, the clinicopathological significance of suppression of FBXW7 expression using small-interfering RNA (siRNA) interference was validated in a GIST cell line.
Materials and methods
Patients
A total of 182 clinical GIST samples were obtained from patients who underwent complete resection at Kumamoto University Hospital, National Hospital Organization Kumamoto Medical Center and Saiseikai Kumamoto Hospital between 2008 and 2018. Ethical approval was obtained from Kumamoto University. Written informed consent was obtained from all patients. GIST was diagnosed by positive staining of c-KIT or CD34 in all resected clinical samples. These cases were divided into four groups according to the frequency of recurrence (very low, low, intermediate and high risk) based on the modified Fletcher classification [19, 20]. The median follow-up period was 741 days. The high-risk patients received imatinib for 3 years as adjuvant therapy after surgery starting in 2011.
Cell line
The human GIST cell line, GIST-T1, was purchased from Cosmo Bio, Co., Ltd. (Tokyo, Japan). GIST-T1 cells were cultured in Dulbecco’s modified Eagle’s medium (Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal bovine serum, at 37 °C under 5% CO2 for at least 1 month.
Immunohistochemical staining of FBXW7
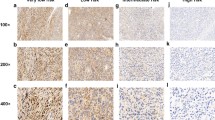
Paraffin-embedded tissues obtained from the GIST patients were cut at a thickness of 4 µm, deparaffinized in xylene, rehydrated in ethanol, and soaked in distilled water. Sample processing and immunohistochemical (IHC) procedures were performed as described below. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide. The sections were incubated with antibodies targeting FBXW7 (1:200; clone 3D1; Abnova, Taipei, Taiwan), c-myc (1:200; ab32072; Abcam, Cambridge, UK), phosphorylated c-myc (1:200; ab51156; Abcam, Cambridge, UK), and Notch 1 (1:100; D1E11; Cell Signaling Technology, Danvers, MA, USA), and detection was performed using enzymatic avidin–biotin complex 3,3′-diaminobenzidine staining (VECTASTAIN Elite ABC mouse and rabbit IgG kit; VEC PK-6101, 6102). FBXW7 expression was scored in tumor cells by three observers according to the immunohistochemical staining intensity (0, no; 1, weak; 2, moderate; or 3, strong staining) and the proportion of stained cells (0–100%) (Fig. 1). FBXW7 expression was divided into the high and low expression groups according to the median staining score (150 points).
Immunohistochemical analysis of FBXW7 expression in GIST-T1 cells. a Immunohistochemical staining intensity score for FBXW7 (0, no; 1, mild; 2, moderate; and 3, strong staining). Magnification ×200. b Two representative cases. Left panel shows a case of high FBXW7 protein expression with low c-myc and Notch 1 expression (low-risk GIST). Right panel shows a case of low FBXW7 protein expression with high c-myc and Notch 1 expression (high-risk GIST). Magnification ×100
Suppression of FBXW7 expression by siRNAs
FBXW7 expression was transiently downregulated using predesigned Silencer Select siRNAs directed against FBXW7 (siFBXW7 #1: s224356; siFBXW7 #2: s224357; Thermo Fisher Scientific, Waltham, MA, USA). A nontargeting siRNA was used as a negative control. GIST-T1 cells were transfected with the siRNAs for 48 h using Lipofectamine RNAiMAX (Thermo Fisher Scientific). At 24 h after transfection, cells were harvested and subjected to Western blotting and quantitative reverse-transcription polymerase chain reaction (qRT-PCR).
qRT-PCR
RNA was isolated from the cultured cells using the RNeasy kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocol. mRNA expression levels were determined by qRT-PCR using TaqMan probes (universal probe #78 for FBXW7; Roche, Basel, Switzerland), and the values were normalized to those of β-actin. All qRT-PCRs were run on the LightCycler 480 System II (Roche). The sequences of the FBXW7 mRNA primers were as follows: forward 5′-ATTGGCAATGAGCGGTTC-3′ and reverse 5′-CGTGGATGCCACAGGACT-3′. β-actin was an internal control using the following primers: forward 5′-TTGGTATCGTGGAAGGACTCTA-3′ and reverse 5′-TGTCATATTTGGCAGGTT-3′. All qRT-PCR experiments were performed in triplicate and the data are presented as mean ± standard error.
Western blot analysis
GIST-T1 cell cultures in 10-cm dishes or 6-well plates were washed twice in PBS and lysed in extraction buffer (Thermo Fisher Scientific) and RIPA lysis buffer supplemented with a protease inhibitor cocktail (Thermo Fisher Scientific). Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with 5% low-fat dry milk in TBST (25 mM Tris [pH 7.4], 125 mM NaCl, 0.4% Tween) and then incubated with primary antibodies targeting c-myc (1:500; Abcam, ab32072), Notch 1 (1:1000; Cell Signaling Technology), and β-actin (1:1000; 13E5; Cell Signaling Technology), which were diluted in TBST at 4 °C overnight for Western blotting. Signals were detected by incubation with mouse and rabbit (1:5000) secondary antibodies at room temperature for 1 h using the ECL Detection System (GE Healthcare, Little Chalfont, UK).
Cytotoxicity assay
Cytotoxicity was evaluated by measuring cell viability using the Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan), a sensitive colorimetric cytotoxicity assay. The amount of formazan dye generated by dehydrogenase activity in cells is directly proportional to the number of living cells. GIST-T1 cells were seeded in 96-well plates (3000/well) in growth medium (100 µl) and incubated at 37 °C under a 5% CO2 atmosphere for 24 h. The cells were then transfected with the FBXW7 siRNAs or control siRNA for 24, 48, 72, or 96 h. CCK-8 reagent was added to each well and the plates were further incubated for 1.5 h. The absorbance of the solution was measured at 450 nm.
Invasion assay
Cell invasion was assessed using BioCoat Matrigel invasion chambers (24-well plates, 8 µm pores; BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. siRNA-transfected GIST-T1 cells (2.5 × 104/well) were seeded in the upper chamber and allowed to invade the lower surface of the filter. After 22 h, the cells were fixed with 100% ethanol and stained with hematoxylin and eosin. The number of cells in three randomly selected fields was counted and statistical analyses were performed using two-tailed Student’s t tests.
Migration assay
siRNA-transfected GIST-T1 cells were seeded in 6-well plates at 5.0 × 104/well and subjected to real-time imaging using the Keyence BZ-X700 all-in-one fluorescence microscope equipped with a CO2- and temperature-controlled chamber and time-lapse tracking system (Keyence, Osaka, Japan). Images were taken every 10 min for 24 h and combined using the BZ-X Analyzer (Keyence). Cell migration was analyzed using the video editing analysis software VW-H2MA (Keyence), and the tracking data were processed using Microsoft Excel 2010 (Microsoft, Redmond, WA, USA) to obtain x, y-coordinate plots and distance measurements.
Statistical analysis
All experiments were repeated at least three times. Continuous variables were expressed as medians (range) or mean ± standard deviation as appropriate. Qualitative variables were compared using the chi-squared test and quantitative variables using the Wilcoxon test. The PCR and proliferation assay data were analyzed using paired Student’s t tests. Data were processed using Excel 2010 (Microsoft). All P values < 0.05 were considered statistically significant. All statistical analyses were performed using JMP version 10 (SAS Institute, Cary, NC, USA).
Results
FBXW7 expression and clinicopathological features of the clinical samples
We examined FBXW7 expression in resected GIST samples by immunohistochemical staining. FBXW7 expression was homogeneously localized to cell nuclei in entire tumor tissue. We divided the 182 GIST cases into high and low expression groups according to the median staining intensity score for FBXW7 (Fig. 1a); 98 (53.8%) and 84 (46.2%) cases were classified into the high and low FBXW7 expression groups. The associations between FBXW7 expression and the clinicopathological factors evaluated are shown in Table 1. The proportions of GISTs located in the duodenum, small intestine, and other sites were significantly higher in the low compared with the high FBXW7 expression group (P = 0.008). Tumor size was significantly larger in the low compared with the high FBXW7 expression group (P = 0.0001). There were significantly more high-risk cases according to the modified Fletcher classification in the low FBXW7 expression group (P = 0.0003). Figure 1b shows two representative cases of GISTs exhibiting inverse correlations of FBXW7 with c-myc and Notch 1 protein expression levels. The case with high FBXW7 expression had low c-myc and Notch 1 expression (left panel), whereas the case with low FBXW7 expression had high c-myc and Notch 1 expression (right panel).
The association between FBXW7 expression and prognosis in GIST patients
The survival curves for 10-year recurrence-free survival (RFS) and overall survival (OS) showed a significantly poorer patient prognosis in the low compared with the high FBXW7 expression group (RFS: P = 0.01; OS: P = 0.03) (Fig. 2a, b). Low FBXW7 expression tended to be correlated with a poor prognosis in the high-risk cases (P = 0.10, Fig. 2c). In the intermediate-/high-risk cases, low FBXW7 expression was significantly correlated with a worse RFS (P = 0.03, Fig. 2d). We also examined the association of FBXW7 expression with prognosis in another cohort and found that low FBXW7 expression was significantly correlated with a worse RFS (P = 0.02, Supplemental Figure 1). Furthermore, we evaluated the association of immunohistochemical expression of c-myc, phosphorylated c-myc and Notch 1 in GIST with patient prognosis. We found that the expression of c-myc, phosphorylated c-myc, and Notch 1 is not significantly associated with prognosis (Supplemental Figure 2). In the univariate analysis, FBXW7 expression, modified Fletcher risk classification, tumor location, and tumor size were identified as prognostic factors for 10-year RFS following curative resection. Variables with P < 0.05 in the univariate analyses were selected for multivariate analysis using Cox’s proportional hazards model, which identified FBXW7 expression (odds ratio (OR) 6.78; 95% confidence interval (CI) 1.10–130.8; P = 0.04) as a significant independent factor affecting the 10-year RFS rate following curative resection (Table 2a). In intermediate-/high-risk patients, FBXW7 expression (OR 6.78; 95% CI 1.10–130.8; P = 0.04) was also a significant factor affecting the 10-year RFS rate following curative resection (Table 2b).
Kaplan–Meier analysis of recurrence-free and overall survival. a Recurrence-free survival and b overall survival curves based on FBXW7 expression in all GIST cases (n = 182). c Recurrence-free survival in the high-risk cases (n = 42), d overall survival curves based on FBXW7 expression in the intermediate-/high-risk cases (n = 65). The high and low FBXW7 expression groups are indicated by the unbroken and broken lines, respectively
The effects of suppression of FBXW7 expression in GIST-T1 cells
To clarify the clinicopathological significance of suppressed FBXW7 expression, we examined the biological effects of FBXW7 siRNA transfection in GIST-T1 cells. Suppression of FBXW7 expression in GIST-T1 cells was observed at 24 h after siRNA transfection (P < 0.01, Fig. 3a). Suppression of FBXW7 induced overexpression of its downstream targets c-myc and Notch 1, as confirmed by Western blot analysis (Fig. 3a).
Effect of FBXW7 gene silencing on GIST-T1 cells. a FBXW7 expression in GIST-T1 cells transfected with FBXW7 siRNAs was suppressed compared with cells transfected with the control siRNA, as confirmed by qRT-PCR. Expression of c-myc and Notch 1 was increased by transfection with the FBXW7 siRNAs, as confirmed by Western blotting. b The proliferation rate of GIST-T1 cells transfected with FBXW7 siRNAs (unbroken line) was significantly greater than that of cells transfected with the control siRNA (broken line). c The number of invading cells after 22 h was significantly greater in GIST-T1 cells transfected with the FBXW7 siRNAs compared with the control siRNA. d GIST-T1 cells transfected with the FBXW7 siRNAs, compared with the control siRNA, migrated significantly greater distances within a 24 h period. Photographs were taken every 10 min for 24 h
In the MTT proliferation assay, the number of viable GIST-T1 cells (3000/well) was evaluated after 24 h incubation with siRNA at several time points (0, 24, 48, 72, and 96 h). We found that the proliferation rate was significantly increased in GIST-T1 cells transfected with FBXW7 siRNAs compared with the control siRNA at 96 h after transfection (siFBXW7 #1, P < 0.01; siFBXW7 #2, P < 0.01; Fig. 3b).
The invasion ability of GIST-T1 cells transfected after 24 h incubation with the FBXW7 siRNAs was examined using BioCoat Matrigel chambers. The number of invading cells after 22 h was significantly greater in GIST-T1 cells transfected with the FBXW7 siRNAs compared with the control siRNA (siFBXW7 #1, P < 0.01; siFBXW7 #2, P < 0.01; Fig. 3c). In the migration assays, we measured the total distance traveled by 10 GIST-T1 cells randomly selected within 24 h. GIST-T1 cells transfected with the FBXW7 siRNAs migrated significantly greater distances in a 24-h period (siFBXW7 #1, P < 0.01; siFBXW7 #2, P < 0.01; Fig. 3d).
Discussion
In the current study, we demonstrated that low FBXW7 expression was associated with a poorer prognosis in both the all and intermediate-/high-risk patients with GISTs, respectively (P = 0.01, P = 0.03). Furthermore, FBXW7 expression was a significant factor affecting the 10-year RFS rate in both the all and intermediate-/high-risk patients (P = 0.04). We also investigated how suppression of FBXW7 expression affects cell growth, migration, and invasion in vitro. These results suggest that FBXW7 is a potential biomarker predicting recurrence after curative resection that may allow more accurate assessment of the indications for adjuvant imatinib treatment and prognosis.
In the pre-imatinib era, more than half of patients with GISTs experienced tumor recurrence [21]. The risk of recurrence in patients after complete resection of primary GISTs without adjuvant therapy is associated with mitotic rate, tumor size, and tumor location [22]. According to risk stratification based on tumor size, mitotic rate, tumor location, and tumor rupture, adjuvant imatinib therapy for high-risk cases is widely accepted worldwide [4, 5]. Although adjuvant therapy using imatinib for high-risk patients improves prognosis, some patients do not benefit from therapy due to toxicity. Furthermore, it remains unclear whether intermediate-risk patients with GISTs are candidates for adjuvant therapy [23]. Therefore, a more precise predictive marker is needed to select the patients who will receive significant benefit from adjuvant therapy. Although more molecular gene alterations have been identified in GISTs, most are not acceptable as predictive markers.
Recent reports showed that cyclin H and p16 are indicators of high-risk GIST [24, 25]. Most recently, Ihle et al. [11] revealed that expression of cell-cycle regulators, such as cyclin D1, CDK4, p21, and p16, was associated with recurrence in 400 high-risk patients with GISTs. These results suggest that genes regulating cell cycle are promising markers predictive of the risk of recurrence after curative resection. In this study, we focused on FBXW7, a regulator of the cell cycle and critical tumor suppressor gene in human cancers. In hepatocellular carcinoma and colorectal cancer, a low level of FBXW7 expression, which affects c-myc and Notch 1 expression, is significantly correlated with cell proliferation, invasion, and migration and poor prognosis [17, 26]. Our in vitro study demonstrated that FBXW7 gene alterations can regulate cell proliferation, invasion, and migration via c-myc and Notch 1 in GIST-T1 cells, which is consistent with several previous reports. Therefore, our results suggest that the malignant behavior of GIST-T1 cells expressing low levels of FBXW7 results in a poorer clinical prognosis. Especially, we found that FBXW7 expression was a significant factor affecting RFS in intermediate-/high-risk patients (Fig. 2d; Table 2b), whereas the expression of c-myc, phosphorylated c-myc, and Notch 1 is not significantly associated with prognosis. This finding suggests that FBXW7 is more useful prognostic marker for GIST because FBXW7 can regulate the stability of multiple oncoproteins, including c-myc, and Notch 1.
Our study has several limitations. First, mutations in FBXW7 have been reported in several types of cancers [27]. Despite the high frequency of FBXW7 mutations observed in other cancers, the FBXW7 mutations associated with GISTs are unknown. Therefore, mutation analyses of FBXW7 in GISTs are needed in future studies. Second, the molecular mechanism of FBXW7 suppression in GISTs remains unclear. We speculate that epigenetic transcriptional regulation [28, 29], translational regulation by non-coding RNAs [18, 30], and copy number loss [17, 31] of FBXW7 are involved, as reported previously. Further studies are needed to identify the mechanism of FBXW7 regulation in GISTs. Despite these limitations, our results have several clinical implications. Although imatinib as adjuvant therapy definitively contributes to improve the prognosis for high-risk GIST, it is possible that the evaluation of FBXW7 in addition to conventional risk stratification can help us to select the patients who should receive adjuvant therapy, especially in intermediate-risk group. This molecular analysis along with clinical risk stratification will help us to guide the personalized therapy for advanced GIST.
In conclusion, we demonstrated by multivariate analysis that FBXW7 expression in GISTs is an independent predictive marker of RFS following surgery. FBXW7 may be a reliable marker to identify patients who will benefit from adjuvant therapy better than conventional risk stratification in a clinical setting.
References
Soreide K, Sandvik OM, Soreide JA, Giljaca V, Jureckova A, Bulusu VR. Global epidemiology of gastrointestinal stromal tumours (GIST): a systematic review of population-based cohort studies. Cancer Epidemiol. 2016;40:39–46.
Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80.
Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, et al. Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–7.
Casali PG, Abecassis N, Bauer S, Biagini R, Bielack S, Bonvalot S, et al. Gastrointestinal stromal tumours: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):v68–iv78.
von Mehren M, Randall RL, Benjamin RS, Boles S, Bui MM, Ganjoo KN, et al. Soft tissue sarcoma, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16:536–63.
DeMatteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231:51–8.
Joensuu H, Eriksson M, Sundby Hall K, Hartmann JT, Pink D, Schutte J, et al. One vs three years of adjuvant imatinib for operable gastrointestinal stromal tumor: a randomized trial. JAMA. 2012;307:1265–72.
Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Int J Surg Pathol. 2002;10:81–9.
Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol. 2002;33:459–65.
Joensuu H, Vehtari A, Riihimaki J, Nishida T, Steigen SE, Brabec P, et al. Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol. 2012;13:265–74.
Ihle MA, Huss S, Jeske W, Hartmann W, Merkelbach-Bruse S, Schildhaus HU, et al. Expression of cell cycle regulators and frequency of TP53 mutations in high risk gastrointestinal stromal tumors prior to adjuvant imatinib treatment. PLoS One. 2018;13:e0193048.
Bashir T, Pagano M. Aberrant ubiquitin-mediated proteolysis of cell cycle regulatory proteins and oncogenesis. Adv Cancer Res. 2003;88:101–44.
Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–81.
Ishikawa Y, Hosogane M, Okuyama R, Aoyama S, Onoyama I, Nakayama KI, et al. Opposing functions of Fbxw7 in keratinocyte growth, differentiation and skin tumorigenesis mediated through negative regulation of c-Myc and Notch. Oncogene. 2013;32:1921–32.
Ishikawa Y, Onoyama I, Nakayama KI, Nakayama K. Notch-dependent cell cycle arrest and apoptosis in mouse embryonic fibroblasts lacking Fbxw7. Oncogene. 2008;27:6164–74.
Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T, et al. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788–94.
Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T, et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int J Cancer. 2010;126:1828–37.
Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–8.
Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol. 2008;39:1411–9.
Rutkowski P, Bylina E, Wozniak A, Nowecki ZI, Osuch C, Matlok M, et al. Validation of the Joensuu risk criteria for primary resectable gastrointestinal stromal tumour—the impact of tumour rupture on patient outcomes. Eur J Surg Oncol. 2011;37:890–6.
Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466–78.
Dematteo RP, Gold JS, Saran L, Gonen M, Liau KH, Maki RG, et al. Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST). Cancer. 2008;112:608–15.
Fu Y, Hao H, Guo L, Yang G, Zhang X. Retrospective analysis of 85 cases of intermediate-risk gastrointestinal stromal tumor. Oncotarget. 2017;8:10136–44.
Schmieder M, Wolf S, Danner B, Stoehr S, Juchems MS, Wuerl P, et al. p16 Expression differentiates high-risk gastrointestinal stromal tumor and predicts poor outcome. Neoplasia. 2008;10:1154–62.
Dorn J, Spatz H, Schmieder M, Barth TFE, Blatz A, Henne-Bruns D, et al. Cyclin H expression is increased in GIST with very-high risk of malignancy. BMC Cancer. 2010;10:350.
Wang X, Zhang J, Zhou L, Sun W, Zheng ZG, Lu P, et al. Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway. Int J Oncol. 2015;47:231–43.
Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–64.
Akhoondi S, Lindstrom L, Widschwendter M, Corcoran M, Bergh J, Spruck C, et al. Inactivation of FBXW7/hCDC4-beta expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010;12:R105.
Kitade S, Onoyama I, Kobayashi H, Yagi H, Yoshida S, Kato M, et al. FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors. Cancer Sci. 2016;107(10):1399–405.
Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int J Cancer. 2015;136:1537–45.
Yokobori T, Mimori K, Iwatsuki M, Ishii H, Tanaka F, Sato T, et al. Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma. Int J Oncol. 2012;41:253–9.
Acknowledgements
The authors thank Dr. Takihiro Kamio, Dr. Reiji Muto and Dr. Toshihiko Murayama for providing clinical samples. The authors also thank Dr. Miyake and Ms. Ogata for their excellent technical assistance.
Funding
This work was supported in part by the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Grant numbers 16K10463 and 16KK0184).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We have no conflicts of interest to declare.
Ethics approval and consent to participate
Ethics approval of this study was granted by the ethics committee at Kumamoto University Hospital (approval number 2212) The study was conducted in accordance with the Declaration of Helsinki principles.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1: Kaplan–Meier analysis of recurrence-free and overall survival in 2nd cohort. (a)
Recurrence-free survival and (b) overall survival curves based on FBXW7 expression in all GIST cases (n = 51). (c) Recurrence-free survival in the high-risk cases (n = 12) and (d) overall survival curves based on FBXW7 expression in the intermediate-/high-risk cases (n = 23). The high and low FBXW7 expression groups are indicated by the unbroken and broken lines, respectively. Supplemental Fig. 2: Kaplan–Meier analysis of recurrence-free and overall survival based on c-myc, p-c-myc and Notch 1 expression. (a) Recurrence-free survival and (b) overall survival curves based on c-myc, p-c-myc and Notch 1 expression in all GIST cases (n = 182). The high and low c-myc, p-c-myc and Notch 1 expression groups are indicated by the unbroken and broken lines, respectively. (PPTX 164 KB)
Rights and permissions
About this article
Cite this article
Koga, Y., Iwatsuki, M., Yamashita, K. et al. The role of FBXW7, a cell-cycle regulator, as a predictive marker of recurrence of gastrointestinal stromal tumors. Gastric Cancer 22, 1100–1108 (2019). https://doi.org/10.1007/s10120-019-00950-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-00950-y