Abstract
Lexical answer type prediction is integral to biomedical question–answering systems. LAT prediction aims to predict the expected answer’s semantic type of a factoid or list-type biomedical question. It also aids in the answer processing stage of a QA system to assign a high score to the most relevant answers. Although considerable research efforts exist for LAT prediction in diverse domains, it remains a challenging biomedical problem. LAT prediction for the biomedical field is a multi-label classification problem, as one biomedical question might have more than one expected answer type. Achieving high performance on this task is challenging as biomedical questions have limited lexical features. One biomedical question must be assigned multiple labels given these limited lexical features. In this paper, we develop a novel feature set (lexical, noun concepts, verb concepts, protein–protein interactions, and biomedical entities) from these lexical features. Using ensemble learning with bagging, we use the label power set transformation technique to classify multi-label. We evaluate the integrity of our proposed methodology on the publicly available multi-label biomedical questions dataset (MLBioMedLAT) and compare it with twelve state-of-the-art multi-label classification algorithms. Our proposed method attains a micro-F1 score of 77%, outperforming the baseline model by 25.5%.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
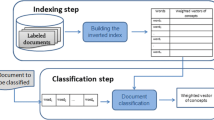
Biomedical knowledge acquisition is an essential task in information retrieval and knowledge management. Professionals and the general public acquire effective assistance to access, understand, and consume complex biomedical concepts [1]. The traditional biomedical question–answering system comprises three components: a question-processing component, a document-processing component, and an answering-processing module, as depicted in Fig. 1. Tremendous developments have been made in biomedical question answering (BQA) in the last two decades, which can be classified into five distinctive approaches: knowledge base, information retrieval, classic, machine reading comprehension, and question entailment [2, 3]. However, despite tremendous growth, BQA still needs to mature and faces many challenges, such as corpora scaling, annotation, lexical answer type prediction, and complex terminology [2, 3].
Lexical answer type (LAT) prediction is a task that aims to predict the type of answer that is best suited for a given question. This helps the question–answering system determine the most appropriate answer based on the LAT. In the open domain, the lexical answer type can be classified as fine-grained or coarse-grained [4], and only one is used for each question. For instance, “When was Barack Obama born?" has only one LAT, which is date. On the other hand, in the biomedical domain, the LAT prediction is a multi-label text classification (MLTC) problem, wherein a single biomedical question can have multiple lexical answer types [5, 6]. For example, the lexical answer type for the biomedical question “Which trinucleotide repeat disorders are affecting the nervous system?" can be disease or syndrome. The development of an effective biomedical question–answering system requires efficient approaches for MLTC to predict multiple lexical answer types for a biomedical question.
MLTC is a supervised machine learning technique in which one document belongs to one or more classes. The previous studies are based on three techniques: problem transformation, adapted algorithms, and ensemble approach [7], as depicted in Fig. 2. Problem transformation approaches include binary relevance, classifier chain, and label power set. The adapted system directly solves the MLTC problems. In this case, multi-label algorithms are used on the data, and no data transformation is required. Finally, ensemble approaches contain a set of multi-label classifiers, such as a classifier chain, to handle multi-label data [8]. Although MLTC has been widely applied in many applications, such as sentiment analysis [9,10,11,12], topic recognition [13, 14], text categorization [15,16,17,18], image classification [19,20,21,22,23], and tag recommendation [7, 8, 24, 25], the work on MLTC for lexical answer type prediction in the biomedical domain remains limited with a low performance affecting the performance of overall question answering system [6, 26].
In this study, we propose a new feature set, inspired by previous research work [6, 26,27,28], to enhance the performance of biomedical lexical answer type prediction in multi-label text classification (MLTC). These features are used in our proposed method, which is based on a label power set with ensemble learning for multi-label classification. In addition, the proposed methodology is rigorously evaluated with other methods for multi-label text classification using a benchmark dataset (MLBioMedLAT [6]). Our study’s primary contributions are summarized as follows:
-
1.
The study introduces discriminative features extracted from the biomedical questions to enhance the MLTC performance.
-
2.
The study proposes a data transformation approach—label power set—with an ensemble learning classifier (Random Forest) for biomedical question’s lexical answer type prediction.
-
3.
The proposed methodology is rigorously analyzed, and a comparison with twelve state-of-the-art models on the benchmark dataset reveals the efficacy of the proposed methodology.
2 Related work
This section presents the work on multi-label text classification (MLTC) in the open and biomedical domains. In the open domain, many previous studies have used multi-label classification techniques on different genres of data, such as movies, legal documents, and medical records [18, 29,30,31,32,33,34,35,36]. The multi-label classification for biomedical questions is a particularly challenging research area because of nature of data [13, 26, 35, 37]. We first briefly present the work in the open domain, followed by the multi-label biomedical question classification work.
2.1 Open-domain MLTC
MLTC techniques have been applied in different domains, including movie genre, law, books, and toxic comments classification [18, 29,30,31,32,33,34,35,36]. For example, Kumar et al. [29] proposed a multi-label classification framework for movie genre classification. They implemented different multi-label classification techniques, including binary relevance and label power set, achieving an 86% F1 score. Similarly, Huang et al. [30] proposed a multi-label classification of users on social media (Twitter) using the KNN algorithm. They proposed an algorithm for multi-label user classification with heterogeneous network and community detection (MLUCHNCD). As a result, MLTC techniques were effective for classification problems, scoring 59% F1 score on the Twitter dataset.
The nature of diverse datasets was also evaluated, and MLTC methods were introduced according to the adaptability of these datasets. A popular method, multi-relation message passing (MrMP), was proposed by Ozmenn et al. [34] for four datasets (Bibtex, Bookmarks, Delicious, and Reuters). This model combined two previously developed models, LaMP and CompGCN, using pulling and pushing relations. The data imbalance problem was also studied by Yang [37] using web pages and newswire articles. They proposed a new model named Hybrid-Siamese convolutional neural network (HSCNN) to address this issue. The proposed model HSCNN outperformed the state-of-the-art systems 77% micro-F1 on entire categories. Another study on the newswire dataset was conducted by Ma [18], who proposed a new architecture named Label Specific Dual Graph Network (LSDG). This model comprised two components: specific document representation and neural network (dual graph) and achieved 97.12% precision on the RCV1 dataset.
Multi-label classification has also been studied for legal documents. Chakidis [38] conducted a study on multi-label classification for the legal domain. The authors released a new dataset consisting of 57k legislative documents. Another research on multi-label classification for books was conducted by Aly [39], proposing a capsule network. Three algorithms, SVM, CNN, and LSTM, were applied to the BGC dataset. Lastly, Pal et al. [40] worked on multi-label text classification for toxic comment dataset. They proposed a new model, MAGNET, based on attendance. This model comprised graph attention network (GAN), correlation matrix, Bi-LSTM, and BERT embedding. The open-domain multilabel text classification literature review indicates that deep learning models such as CNNs, LSTMs, and transformers have shown promising results. In the next section, we briefly cover MLTC for the biomedical domain.
2.2 Biomedical MLTC
In this section, we will primarily focus on text classification in the biomedical domain. Biomedical text can take many forms, including scientific articles, clinical notes, and questions posed by both laypersons and biomedical experts on online forums. Research on MLTC was conducted using benchmark datasets related to COVID-19. Two different approaches were proposed by researchers. Lin et al. [31] utilized a BERT-based ensemble learning model for the COVID benchmark dataset. The model comprised the input layer, multi-head attention layer, and output layer. On the other hand, Chen et al. [33] focused on a multi-label framework for biomedical documents related to COVID-19, specifically using the LitCOVID database on PubMed. They utilized the transformer model named LITMC-BERT and observed the significance of pre-trained deep architectures in achieving better results. Machine learning techniques have been applied to various areas such as clinical notes, biomedical text indexing, and international coding classification (ICD). In a recent study, four machine learning models, including SVM, XGboost, KNN, Random Forest, and a deep learning model called Bi-LSTM, were used to apply multi-label techniques on clinical notes [36]. Another study proposed an end-to-end model named ML-Net [41], which consisted of three modules: document encoding network, label prediction network, and label count prediction network. These models have been applied in biomedical text indexing using MLTC models named CSS and Labelglosses [42]. Multi-label learning has also been used for international coding classification (ICD) using the MIMIC-III dataset [32]. The proposed model achieved a 72.8% F1 score on the MIMIC-III dataset by using 50 candidate clinical notes with prompt-based fine-tuning. The multi-label classification of questions posed by a layman or biomedical experts is essential for effective biomedical question–answering systems [6, 26, 35]. In this regard, one recent study developed a CADEC dataset for multi-label classification of medical forum questions [35] showing their proposed MedBERT superior performance with a macro-F1 score of 0.71. Furthermore, a multi-label biomedical question classification corpus named MLBioMedLAT was developed based on a benchmark question–answering dataset (BioASQ) [6]. In this study, the researchers extracted eight features using the wrapper-based feature selection method. They transformed the multi-label data using copy transformation and label power set transformation and classified the questions using logistic regression, outperforming with a 50% F1 score. Another research on the same benchmark dataset was conducted by Peng et al. [26]. The authors used principal component analysis (PCA) to decrease the dimensionality of features. They achieved a 51.5% F1. However, both these studies have a lower F1 score on the benchmark dataset. A comparison of previous studies in the open and biomedical domain is shown in Table 1. Although there are considerable studies in the medical domain, the work in the biomedical domain for multi-label question classification is limited. Furthermore, there is only one dataset (MLBioMedLAT) which the researchers have used previously to evaluate the performance of their models. The next section presents our proposed multi-label biomedical question classification methodology.
3 Proposed methodology
In this section, we provide details about the proposed methodology. Firstly, a brief introduction to the MLBioMedLAT benchmark dataset is presented. Secondly, we provide details on the preprocessing and feature extraction process. Five features (lexical, noun concepts, verb concepts, biomedical named entity, and protein–protein interaction) were extracted from the questions. The data transformation with the label power set and an ensemble learning-based multi-label classification is presented next. Lastly, we present the proposed methodology evaluation process. The complete process is depicted in Fig. 3.
3.1 MLBioMedLAT: the benchmark dataset
In this study, we use the multi-label Biomedical Lexical Answer Type (MLBioMedLAT) corpus [6]. The corpus contains 780 questions from the fifth year of the BioASQ training dataset [45]. The dataset was annotated with 85 lexical answer types. These types include UMLS semantic types, tmtool, and two additional answer types (choice and quantity) in the BioASQ dataset. The lexical answer type with the most annotated questions (213) from UMLS semantic types is aapp: AminoAcid, Peptide, or Protein. On the other hand, only one question was annotated with the lexical answer type of Protein Mutation. The complete dataset statistics, annotation process, and guidelines are available in the previous study [6].
3.2 Preprocessing and feature extraction
The first step in our methodology is preprocessing the biomedical questions. We remove stop words and punctuation marks from biomedical questions using the neattextFootnote 1 library. Secondly, we analyzed the work on previous studies for MLTC on biomedical question classification [6, 26,27,28] and selected the best-performing features for our proposed methodology. These features include lexical, protein–protein interaction, noun concepts, biomedical entities, and verb concepts. The details of each feature are presented in subsequent sections.
3.2.1 Lexical features
Lexical features are the unigrams from the biomedical questions. These features are extracted after removing the stop words and punctuation marks [6, 26]. For example, the lexical features for the question List signaling molecules (ligands) that interact with the receptor EGF are List, signaling, molecules, ligands, interact, receptor, and EGF.
3.2.2 Protein–protein interaction
The protein–protein Interaction feature is used to find a binary value of either 0 or 1 in biomedical questions [27, 28]. If the question contains protein information, its value is one; otherwise, it is 0. This feature is extracted using GeniaTagger.Footnote 2 For example, the question Which histone modifications are associated with Polycomb group (PcG) protein? contains protein information, so its value will be one. This feature has a significant impact on biomedical text classification.
3.2.3 Noun concepts
There is a strong correlation between noun phrases and biomedical text classification [6, 27]. Therefore, we use noun phrases of biomedical questions to extract the UMLS specialist lexicon. First, we take the noun phrases from biomedical questions using biomedical GeniaTagger. Then, these phrases are passed to MetaMap APIFootnote 3 to get noun concepts. For example, the noun concepts of the question: List protein gel staining methods visualizing the entire protein. The noun concepts from the Metamap API are [inpr], [sbst], [chem], [aapp,rcpt], and [aapp, rcpt]. So, five noun concepts are extracted from this question.
3.2.4 Biomedical entities
Biomedical questions have biomedical entities such as genes, chemicals, viruses, and proteins [6, 27]. We use the biomedical Scispacy Library to extract entities from questions. This library extracts entities of type, such as protein, gene, or simple chemical. For example, biomedical entities pulled from the question: Which thyroid hormone transporter is implicated in thyroid hormone resistance syndrome? The biomedical entities from ScispacyFootnote 4 are: thyroid hormone transporter and thyroid hormone resistance syndrome.
3.2.5 Verb concepts
Verb phrases contribute significantly to the classification of text [6, 27]. We extracted verb phrases from biomedical questions. Again, we used MetaMap API to get verb phrase concepts. For example, the verb concept of the question Which miRNAs could be used as potential biomarkers for epithelial ovarian cancer? The verb concept from the Metamap API is [fndg].
3.3 Label power set data transformation
Label power set is a problem transformation technique for MLTC tasks. It converts MLTC to multi-class problems. It transforms data with all unique label groups in the dataset. As a result, it also captures the label dependency and correlation in the dataset. The transformation procedure in the Label power set is described below. Table 2 shows a small subset of the dataset to explain this transformation.
From Table 3, we can see that example 1,3 and example 2,4 have the same set of labels. Label power set transforms the dataset into a single multi-class classification problem. Table 4 shows this transformation.
3.4 Ensemble learning
Ensemble learning is generally a Meta approach to machine learning that seeks better predictive performance by aggregating the predictions from multiple models. Bagging, boosting, and stacking are the most dominant in the ensemble learning field. We investigated and applied different multi-label classifiers to examine their effect on the predictive performance of the proposed model. One of the ensemble learning methods for classification, regression, and the tasks that acquire a multitude of decision trees for the training time of the dataset is random forests or random decision forests. For multi-label text classification, we applied the random forest model with bagging as the ensemble method and the decision tree as the individual model. We selected the number of rows for random subsets from the training dataset. One hundred decision trees were trained, and one random subset was used to train one decision tree. Each tree predicts the instances in the test dataset independently. The final prediction is made by combining the individual predictions by majority voting for each candidate in the test set. The multi-label classification process is presented in Algorithm 1.
3.5 Performance evaluation
We used example-based evaluation measures for the MLTC task [?]. These measures include Micro-F1 and hamming loss. Micro-F1 is used to measure the aggregated contribution of all classes. The formula of micro-F1 is:
Hamming loss is the average number of errors found in the instance-label pairs, averaged over all the instances, and a lower hamming loss value represents better classification performance. It can be defined as:

Where N is the total number of instances in the dataset, \(|y_i\Delta \ \hat{y}_i|\) is the size of the symmetric difference between the true labels and predicted labels, and L is the number of unique labels in the dataset.
4 Results and discussion
This section presents the experimental results of multi-label biomedical question classification. We used Python’s multi-learnFootnote 5 to experiment with different multi-label algorithms for the MLTC task on the MLBioMedLAT corpus. First, we extracted lexical features, nouns, verbs, biomedical entities, and protein–protein interactions from the biomedical questions. Secondly, we used three transformation techniques: Label power set, Binary Relevance, and Classifier chain. Furthermore, we implemented one adapted algorithm, MLKNN, and an ensemble learning approach, RAKEL. We also compared our methodology with other multi-label classification approaches, including multinomial Naïve Bayes, logistic regression, and deep learning models ClinicalBERT and RoBERTa on the MLBioMedLAT benchmark dataset as shown in Table 5. The experimental results suggest that the Label power set transformation technique with random forest handles the benchmark multi-label data most effectively, achieving a micro-F1 score of 77%. The ClinicalBERT model achieved the lowest score 57.9% Micro-F1. The reason for its low performance was the nature of biomedical questions, which have limited context, thus making it challenging for the deep learning models to perform well.
Table 6 shows the results of our label power set with random forest on different combinations of features. We incrementally add features and observe the effect of each feature on performance metrics. First, we combine lexical features with all other features and only keep the best combination with lexical features. This way, we incrementally added features and keep only the best ones. The table demonstrates that combining all features has the highest micro-F1. The table shows that the micro-F1 score is 75.3% on lexical features. The features of noun and verb concepts increase the evaluation results.
On the other hand, the feature of the biomedical entity had no impact on the results. Furthermore, the feature protein–protein interaction decreases the evaluation performance. Figure 4 describes the performance results of the proposed methodology. Label power set with random forest classifier shows the highest evaluation results in micro-F1.
Hamming loss for classifying biomedical questions by multi-labels is shown in Fig. 5. Label power set with random forest classifier has minimum hamming loss score [6, 26]. As illustrated in Table 7, our proposed methodology is compared to a state-of-the-art biomedical multi-label question classification model.
Our multi-label classification of biomedical questions uses a combination of different features compared with the previous work. These semantic features improved the performance of the LAT prediction task. We also compared the contribution of individual features in the LAT prediction task. Figure 6 shows that the noun concept feature is vital in improving the model’s performance.
Our work outperforms with a margin of 25.5%, as shown in Table 7. Previously, there was a 51.5% micro-F1 score on the MLBioMedLAT corpus. We improved the micro-F1 score to 77%.
5 Conclusion
In this paper, we proposed a multi-label lexical answer type (LAT) prediction method using novel semantic features and a label power set transformation approach. Furthermore, we used bagging for ensemble learning as a classification approach. The previous work on multi-label biomedical question classification had deficient performance (51.5% micro-f1 score) on this task. We proposed novel semantic features for biomedical questions, which included lexical, noun concepts, verb concepts, protein–protein interaction, and named entities. Furthermore, we investigated the impact of different types of features on the performance of the LAT prediction task. To evaluate the performance of our proposed methodology, we used a benchmark corpus (MLBioMedLAT). Our proposed method overshadowed the performance of twelve state-of-the-art multi-label classification datasets on the benchmark dataset. Finally, we compared our proposed methodology with the baseline study, and it outperformed the previous research by a margin of 25.5%, attaining a micro-F1 score of 77%. We plan to explore more advanced feature engineering methods and automatic feature representation techniques in the future.
References
Shortliffe EH, Chiang MF (2021) Biomedical data: their acquisition, storage, and use. Biomedical informatics: computer applications in health care and biomedicine. Springer, Cham, pp 45–75
Jin Q, Yuan Z, Xiong G, Yu Q, Ying H, Tan C, Chen M, Huang S, Liu X, Yu S (2022) Biomedical question answering: a survey of approaches and challenges. ACM Comput Surv (CSUR) 55(2):1–36
Antoniou C, Bassiliades N (2022) A survey on semantic question answering systems. Knowl Eng Rev 37:2
Li X, Roth D (2002) Learning question classifiers. In: COLING 2002: the 19th international conference on computational Linguistics
Neves M, Kraus M (2016) Biomedlat corpus: annotation of the lexical answer type for biomedical questions. In: Proceedings of the open knowledge base and question answering workshop (OKBQA 2016), pp 49–58
Wasim M, Asim MN, Khan MUG, Mahmood W (2019) Multi-label biomedical question classification for lexical answer type prediction. J Biomed Inform 93:103143
Izadi M, Heydarnoori A, Gousios G (2021) Topic recommendation for software repositories using multi-label classification algorithms. Empir Softw Eng 26(5):93
Prajapati P, Thakkar A (2022) Performance improvement of extreme multi-label classification using k-way tree construction with parallel clustering algorithm. J King Saud Univ Comput Inf Sci 34(8):6354–6364
Kumar JA, Trueman TE, Cambria E (2022) Gender-based multi-aspect sentiment detection using multilabel learning. Inf Sci 606:453–468
Shi W, Li F, Li J, Fei H, Ji D (2022) Effective token graph modeling using a novel labeling strategy for structured sentiment analysis. In: Proceedings of the 60th annual meeting of the association for computational Linguistics, Vol. 1. Long Papers, pp 4232–4241
Jain PK, Pamula R, Yekun EA (2022) A multi-label ensemble predicting model to service recommendation from social media contents. J Supercomput 78(4):5203–5220
Deniz E, Erbay H, Coşar M (2022) Multi-label classification of e-commerce customer reviews via machine learning. Axioms 11(9):436
Chen Z, Ren J (2021) Multi-label text classification with latent word-wise label information. Appl Intell 51(2):966–979
Javeed A (2023) Hawk: an industrial-strength multi-label document classifier. arXiv preprint arXiv:2301.06057
Balamurugan V, Vedanarayanan V, Sahaya Anselin Nisha A, Narmadha R, Amirthalakshmi T (2022) Multi-label text categorization using error-correcting output coding with weighted probability. Int J Eng 35(8):1516–1523
Lee J, Yu I, Park J, Kim D-W (2019) Memetic feature selection for multilabel text categorization using label frequency difference. Inf Sci 485:263–280
Vaissnave V, Deepalakshmi P (2022) A keyword-based multi-label text categorization in the Indian legal domain using bi-lstm. Soft computing theories and applications proceedings of SoCTA. Springer, Cham, pp 213–227
Ma Q, Yuan C, Zhou W, Hu S (2021) Label-specific dual graph neural network for multi-label text classification. In: Zong C, Xia F, Li W, Navigli R. (eds.) Proceedings of the 59th annual meeting of the association for computational Linguistics and the 11th international joint conference on natural language processing, Vol. 1. Long Papers, pp 3855–3864. Association for Computational Linguistics, Onlinehttps://doi.org/10.18653/v1/2021.acl-long.298.https://aclanthology.org/2021.acl-long.298
Pu T, Sun M, Wu H, Chen T, Tian L, Lin L (2023) Semantic representation and dependency learning for multi-label image recognition. Neurocomputing 526:121–130
Abdel-Khalek S, Algarni M, Mansour RF, Gupta D, Ilayaraja M (2021) Quantum neural network-based multilabel image classification in high-resolution unmanned aerial vehicle imagery. Soft Comput 1–12
Xu J, Tian H, Wang Z, Wang Y, Kang W, Chen F (2020) Joint input and output space learning for multi-label image classification. IEEE Trans Multimedia 23:1696–1707
Coulibaly S, Kamsu-Foguem B, Kamissoko D, Traore D (2022) Deep convolution neural network sharing for the multi-label images classification. Mach Learn Appl 10:100422
Liang J, Xu F, Yu S (2022) A multi-scale semantic attention representation for multi-label image recognition with graph networks. Neurocomputing 491:14–23
Bogatinovski J, Todorovski L, Džeroski S, Kocev D (2022) Comprehensive comparative study of multi-label classification methods. Expert Syst Appl 203:117215
Erlich A, Dantas SG, Bagozzi BE, Berliner D, Palmer-Rubin B (2022) Multi-label prediction for political text-as-data. Polit Anal 30(4):463–480
Peng K, Rong W, Li C, Hu J, Xiong Z (2020) Weight aware feature enriched biomedical lexical answer type prediction. In: Neural information processing: 27th international conference, ICONIP 2020, Bangkok, Thailand, 23–27 Nov 2020, Proceedings, Part III 27. Springer, pp 63–75
Muzaffar AW, Azam F, Qamar U (2015) A relation extraction framework for biomedical text using hybrid feature set. Comput Math Methods Med 2015:910423
Ahmed M, Islam J, Samee MR, Mercer RE (2019) Identifying protein-protein interaction using tree lstm and structured attention. In: 2019 IEEE 13th international conference on semantic computing (ICSC). IEEE, pp 224–231
Kumar S, Kumar N, Dev A, Naorem S (2023) Movie genre classification using binary relevance, label powerset, and machine learning classifiers. Multimedia Tools Appl 82(1):945–968
Huang A, Xu R, Chen Y, Guo M (2023) Research on multi-label user classification of social media based on ml-knn algorithm. Technol Forecasting Soc Change 188:122271
Lin S-J, Yeh W-C, Chiu Y-W, Chang Y-C, Hsu M-H, Chen Y-S, Hsu W-L (2022) A bert-based ensemble learning approach for the biocreative vii challenges: full-text chemical identification and multi-label classification in pubmed articles. Database 2022:056
Yang Z, Wang S, Rawat BPS, Mitra A, Yu H (2022) Knowledge injected prompt based fine-tuning for multi-label few-shot icd coding. In: Proceedings of the conference on empirical methods in natural language processing. Conference on empirical methods in natural language processing, vol. 2022. NIH Public Access, p 1767
Chen Q, Du J, Allot A, Lu Z (2022) Litmc-bert: transformer-based multi-label classification of biomedical literature with an application on covid-19 literature curation. IEEE/ACM Trans Comput Biol Bioinform 19(5):2584–2595
Ozmen M, Zhang H, Wang P, Coates M (2022) Multi-relation message passing for multi-label text classification. In: ICASSP 2022-2022 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 3583–3587
Roy S, Chakraborty S, Mandal A, Balde G, Sharma P, Natarajan A, Khosla M, Sural S, Ganguly N(2021) Knowledge-aware neural networks for medical forum question classification. In: Proceedings of the 30th acm international conference on information & knowledge management, pp 3398–3402
Stemerman R, Arguello J, Brice J, Krishnamurthy A, Houston M, Kitzmiller R (2021) Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open 4(3):069
Yang W, Li J, Fukumoto F, Ye Y (2020) Hscnn: a hybrid-siamese convolutional neural network for extremely imbalanced multi-label text classification. In: Proceedings of the 2020 conference on empirical methods in natural language processing (EMNLP), pp 6716–6722
Chalkidis I, Fergadiotis E, Malakasiotis P, Androutsopoulos I (2019) Large-scale multi-label text classification on eu legislation. In: Proceedings of the 57th annual meeting of the association for computational Linguistics, pp 6314–6322
Aly R, Remus S, Biemann C (2019) Hierarchical multi-label classification of text with capsule networks. In: Proceedings of the 57th annual meeting of the association for computational Linguistics: student research workshop, pp 323–330
Pal A, Selvakumar M, Sankarasubbu M (2020) Multi-label text classification using attention-based graph neural network. arXiv preprint arXiv:2003.11644
Du J, Chen Q, Peng Y, Xiang Y, Tao C, Lu Z (2019) Ml-net: multi-label classification of biomedical texts with deep neural networks. J Am Med Inform Assoc 26(11):1279–1285
Zhang Y, Li X, Liu Y, Li A, Yang X, Tang X (2023) A multilabel text classifier of cancer literature at the publication level: methods study of medical text classification. JMIR Med Inform 11(1):44892
Ma Y, Liu X, Zhao L, Liang Y, Zhang P, Jin B (2022) Hybrid embedding-based text representation for hierarchical multi-label text classification. Expert Syst Appl 187:115905
Wang R, Ridley R, Qu W, Dai X (2021) A novel reasoning mechanism for multi-label text classification. Inf Process Manag 58(2):102441
Nentidis A, Bougiatiotis K, Krithara A, Paliouras G, Kakadiaris I (2017) Results of the fifth edition of the bioasq challenge. In: BioNLP 2017, pp 48–57
Acknowledgements
This work is funded by FCT/MEC through national funds and, when applicable, co-funded by the FEDER-PT2020 partnership agreement under the project UIDB/50008/2020.
Funding
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hussain, F.G., Wasim, M., Cheema, S.M. et al. Semantic features analysis for biomedical lexical answer type prediction using ensemble learning approach. Knowl Inf Syst 66, 5003–5019 (2024). https://doi.org/10.1007/s10115-024-02113-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-024-02113-7