Abstract
This paper contributes multivariate versions of seven commonly used elastic similarity and distance measures for time series data analytics. Elastic similarity and distance measures can compensate for misalignments in the time axis of time series data. We adapt two existing strategies used in a multivariate version of the well-known Dynamic Time Warping (DTW), namely, Independent and Dependent DTW, to these seven measures. While these measures can be applied to various time series analysis tasks, we demonstrate their utility on multivariate time series classification using the nearest neighbor classifier. On 23 well-known datasets, we demonstrate that each of the measures but one achieves the highest accuracy relative to others on at least one dataset, supporting the value of developing a suite of multivariate similarity and distance measures. We also demonstrate that there are datasets for which either the dependent versions of all measures are more accurate than their independent counterparts or vice versa. In addition, we also construct a nearest neighbor-based ensemble of the measures and show that it is competitive to other state-of-the-art single-strategy multivariate time series classifiers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Elastic similarity and distance measures, such as the well-known Dynamic Time Warping (DTW) [1], are a key tool in many forms of time series analytics. Elastic similarity and distance measures can align temporal misalignments between two series while computing the similarity or distance between them. Examples of their application include clustering [2,3,4], classification [5, 6], anomaly detection [7, 8], indexing [9], subsequence search [10] and segmentation [11].
While numerous elastic similarity and distance measures have been developed [6, 12, 13], most of these measures have been defined only for univariate time series. One elastic measure that has previously been extended to the multivariate case is DTW [14]. That work identified two key strategies for such extension. The independent strategy applies the univariate measure to each dimension and then sums the resulting distances. The dependent strategy treats the multivariate series as a single series in which each time step has a single multidimensional point. DTW is then applied using Euclidean distances between the multidimensional points of the two series.
This paper extends seven further key univariate similarity and distance measures, presented in Table 1, to the multivariate case. We choose these seven specific measures because our research is largely focused on classification and these measures have been used in many well-known univariate similarity and distance-based classifiers such as Elastic Ensemble [5], Proximity Forest [15], and two state-of-the-art univariate time series classifiers HIVE-COTE 1.0 [16] and TS-CHIEF [17].
This extension is important, because many real-world time series are multidimensional. For some measures, it is straightforward, but non-trivial for three measures, LCSS, MSM, and TWE. We show that each measure except one provides more accurate nearest neighbor classification than any alternative for at least one dataset. This demonstrates the importance of having a range of multivariate elastic measures.
It has been shown that the dependent and independent strategies each outperformed the other on some tasks when applied to DTW [14]. One of this paper’s significant outcomes is to demonstrate that there are some tasks for which the independent strategy is superior across all measures and others for which the dependent strategy is better. This establishes a fundamental relationship between the two strategies and different tasks, countering the possibility that differing performance for the two strategies when applied to DTW might have been coincidental.
We further illustrate the value of multiple measures by developing a multivariate version of the Elastic Ensemble [5]. We demonstrate that this ensemble of nearest neighbor classifiers using all multivariate measures provides accuracy competitive with the state-of-the-art single strategies in multivariate time series classification.
We organize the rest of the paper as follows. Section 2 presents key definitions and a brief review of existing methods. Section 3 describes our new multivariate similarity and distance measures. Section 4 presents multivariate time series classification experiments on the UEA multivariate time series archive, and includes discussion of the implications of the results. Finally, we draw conclusions in Sect. 5, with suggestions for future work.
2 Related work
2.1 Definitions
We here present key notations and definitions.
A time series T of length L is an ordered sequence of L time-value pairs \( T = \langle (t_1, \varvec{x}_1), \ldots , (t_L, \varvec{x}_L) \rangle \), where \(t_i\) is the timestamp at sequence index i, \(i \in \{1, \ldots , L\}\), and \(\varvec{x}_i\) is a D-dimensional point representing observations of D real-valued variables or features at timestamp \(t_i\). Each time point \(\varvec{x}_i\in {\mathbb {R}}^D\) is defined by \(\{x^1_i, \ldots ,x^d_i, \ldots ,x^D_i \}\).
Usually, timestamps \(t_i\) are assumed to be equidistant, and thus omitted, which results in a simpler representation where \(T~=~\langle \varvec{x}_1,\ldots , \varvec{x}_L \rangle \).
A univariate (or single-dimensional) time series is a special case where a single variable is observed (\(D=1\)). Therefore, \(\varvec{x}_i\) is a scalar, and consequently, \(T = \langle x_1, \ldots , x_L\rangle \).
A labeled time series dataset \({\mathcal {S}}\) consists of n labeled time series indexed by k, where \(k \in \{1, \ldots , n\}\). Each time series \(T_k\) in \({\mathcal {S}}\) is associated with a label \(y_k\in \{1,\ldots ,c\}\), where c is the number of classes.
A similarity measure computes a real value that quantifies similarity between two sets of values. A distance measure computes a real value that quantifies dissimilarity between two sets of values. For time series Q and C, a similarity or distance measure M is defined as
A measure M is a metric if it has the following properties:
-
1.
Non-negativity: \(M(Q, C) \ge 0\),
-
2.
Identity: \(M(Q, C) = 0\), if and only if \(Q = C\),
-
3.
Symmetry: \(M(Q, C) = M(C, Q)\),
-
4.
Triangle Inequality: \(M(Q, C) \le M(Q, T) + M(T, C)\) for any time series Q, C and T.
In a Time Series Classification (TSC) task, a time series classifier is trained on a labeled time series dataset, and then used to predict labels of unlabeled time series. The classifier is a predictive mapping function that maps from the space of input variables to discrete class labels.
In this paper, to perform TSC tasks, we use 1-nearest neighbor (1-NN) classifiers, which use time series-specific similarity and distance measures to compute the nearest neighbors between each time series.
2.2 Univariate TSC
A comprehensive review of the most common univariate TSC methods developed prior to 2017 can be found in [6]. Here we summarize key univariate TSC methods following a widely used categorization as follows:
-
Similarity and distance-based methods compare whole time series using similarity and distance measures, usually in conjunction with 1-NN classifiers. Particularly, 1-NN with DTW [1, 26] was long considered as the de facto standard for univariate TSC. More accurate similarity and distance-based methods combine multiple measures, including 1-NN-based ensemble Elastic Ensemble (EE) [5], and tree-based ensemble Proximity Forest (PF) [15]. In Sect. 3 we will explore more details of several similarity and distance measures used in TSC.
-
Interval-based methods use summary statistics relating to subseries in conjunction with location information as discriminatory features. Examples include Time Series Forest (TSF) [27], Random Interval Spectral Ensemble (RISE) [16], Canonical Interval Forest (CIF) [28] and Diverse Representation CIF (DrCIF) [28]. Currently, DrCIF is the most accurate classifier in this category [29].
-
Shapelet-based methods extract or learn a set of discriminative subseries for each class which are then used as search keys for the particular classes. The presence, absence or distance of a shapelet is used as discriminative information for classification. Examples include Shapelet Transform (ST) [30] and Generalized Random Shapelet Forest (gRSF) [31], and a time contracted version of ST called Shapelet Transform Classifier (STC) [32].
-
Dictionary-based methods transform time series into a bag-of-word model. The series is either discretized in time domain such as in Bag of Patterns (BoP) [33] or it is transformed into the frequency domain such as in Bag-of-SFA-Symbols (BOSS) [34], and Word eXtrAction for time SEries cLassification (WEASEL) [35] and Temporal Dictionary Ensemble (TDE) [36]. Currently, TDE is the most accurate classifier in this category [29].
-
Kernel-based methods transform the time series using a transformation function and then use a general purpose classifier. A notable example is RandOm Convolutional KErnel Transform (ROCKET) [37] which uses random convolutions to transform the data, and then uses logistic regression for classification.
-
Deep-learning methods can be divided into two main types of architectures: (1) based on recurrent neural networks [38], or (2) based on temporal convolutions, such as Residual Neural Network (ResNet) [39] and InceptionTime [40]. A recent review of deep learning methods shows that architectures that use temporal convolutions show higher accuracy [41].
-
Combinations of Methods combine multiple methods to form ensembles. Examples include HIVE-COTE (Hierarchical Vote Collective of Transformation-based Ensembles) [16], which ensembles EE, ST, RISE and BOSS, and TS-CHIEF (Time Series Combination of Heterogeneous and Integrated Embeddings Forest) [17], which is a tree-based ensemble where the tree nodes use similarity, distance, dictionary or interval-based splitters.
InceptionTime, TS-CHIEF, ROCKET and HIVE-COTE have been identified to be state-of-the-art classifiers for TSC [29]. The latest versions of HIVE-COTE, which do not include EE, called HIVE-COTE 1.0 [32] and HIVE-COTE 2.0 [29], significantly improved the speed of the original version [16]. While benchmarking on the UCR univariate TSC archive places HIVE-COTE 1.0 behind ROCKET and TS-CHIEF on accuracy [32], recent benchmarking [29] places HIVE-COTE 2.0 ahead on accuracy relative to all alternatives.
2.3 Multivariate TSC
Research into multivariate TSC has lagged behind univariate research. A recent paper [42] reviews several methods used for multivariate TSC and compares their performance on the UEA multivariate time series archive [43]. Here, we present a short summary of multivariate methods:
-
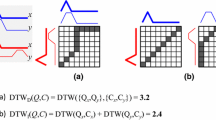
Multivariate similarity and distance measures can be used with 1-nearest neighbor for classification. DTW has previously been extended to the multivariate case using two key strategies [14]. The independent strategy applies the univariate measure to each dimension and then sums the resulting distances. The dependent strategy treats each time step as a multidimensional point. DTW is then applied on the Euclidean distances between these multidimensional points. Figure 1 illustrates these approaches, and we present definitions in Sect. 3. In addition, Shokoohi et al. [14] present an adaptive approach to select between independent or dependent DTW based on the performance on the dataset. Since this adaptive approach falls back to either the independent or dependent version based on the performance, we only studied the independent and dependent strategies when experimenting with single measures. However, in Sect. 3.7 we present an ensemble based on our own version of an adaptive approach. We discuss its results in Sect. 4.4.
-
Interval-based methods include RISE [16], TSF [27], and the recently introduced CIF [28] and its extension DrCIF [29]. They extract intervals from each dimension separately. CIF and DrCIF have shown promising results for multivariate classification [42].
-
Shapelet-based methods include gRSF [31] and time contracted Shapelet Transform (STC) [32]. According to the review [42], STC is the current most accurate multivariate method that uses shapelets [42].
-
Dictionary-based methods include WEASEL with a Multivariate Unsupervised Symbols and dErivatives (MUSE) (a.k.a WEASEL+MUSE, or simply MUSE) [35] and time contracted Bag-of-SFA-Symbols (CBOSS) [44].
-
Kernel-based methods include an extension of ROCKET to the multivariate case, implemented in the sktime library [45]. It combines information from multiple dimensions using small subsets of dimensions.
-
Combinations of Methods include a multivariate version of HIVE-COTE 1.0 which combines STC, TSF, CBOSS, and RISE [32], applying each constituent algorithm to each dimension separately.
-
Deep-learning-based methods that directly support multivariate series include Time Series Attentional Prototype Network (TapNet) [46], ResNet [41] and InceptionTime [40].
To benchmark key algorithms covered in the review, Ruiz et al. compared 12 classifiers on 20 UEA multivariate datasets with equal length that completed in a reasonable time. They found that the most accurate multivariate TSC algorithms are ROCKET, InceptionTime, MUSE, CIF, HIVE-COTE and MrSEQL in that order [42, Fig. 12a].
3 Similarity and distance measures
In this section, we present the proposed similarity and distance measures. For this study, we extend to the multivariate case the set of univariate similarity and distance measures used in EE and PF (and thus TS-CHIEF and some versions of HIVE-COTE).
The independent strategy proposed by [14] simply sums over the results of applying DTW separately to each dimension. For completeness, we propose to extend this idea to allowing any p-norm. In this case, the previous approach extends directly to any univariate measure as follows.
Definition 1
Independent Measures For any univariate measure \(m(Q^1,C^1)\rightarrow {\mathbb {R}}\) and multivariate series Q and C, an independent multivariate extension of m is defined by
We compute the distance between Q and C separately for each dimension, and then take the p-norm of the results. Here, \(Q^d\) (or \(C^d\)) represents the univariate time series of dimension d such that \(Q^d=<q_1^d,\ldots ,q_L^d>\) (or \(C^d=<c_1^d,\ldots ,c_L^d>\)). The parameter p is set to 1 in Shokoohi-Yekta et al. [14].
For consistency with previous work, we assume a 1-norm unless otherwise specified. For ease of comprehension, we indicate an independent extension of a univariate measure by adding the subscript I. Hence, \(DTW_I(Q,C)=Ind(DTW, Q, C, 1)\), \(WDTW_I(Q,C)=Ind(WDTW, Q, C, 1)\) and so forth.
However, in most cases it requires more than such a simple formulation to derive a dependent extension, and hence we below introduce each of the univariate measures together with our proposed dependent variant.
3.1 Lp distance (Lp)
3.1.1 Univariate Lp distance
The simplest way to calculate distance between two time series is to use Lp distance, also known as the Minkowski distance.
Let us denote by Q and C two univariate (\(D = 1\)) time series of length L where \(q_i\) and \(c_i\) are scalar values at time point i from the two time series. Equation 3 formulates the Lp distance between Q and C.
The parameter p is the order of the distance. The \(L_1\) (Manhattan distance) and \(L_2\) (Euclidean distance) distances are widely used.
3.1.2 Multivariate Lp distance
We here show that Independent Lp distance (\(Lp_{I}\)) and Dependent Lp distance (\(Lp_{D}\)) are identical for a given value of the parameter p.
Definition 2
Independent Lp Distance (\(Lp_I\)) In this case, we simply compute the Lp distance between Q and C separately for each dimension, and then take the p-norm of the results.
Definition 3
Dependent Lp Distance (\(Lp_D\)) In this case, we compute the Lp distance between each multidimensional point \(\mathbf {q_{i}}\in {\mathbb {R}}^D\) and \(\mathbf {c_{i}}\in {\mathbb {R}}^D\), and take the p-norm of the results.
Consequently both the independent and the dependent versions of the non-elastic Lp distance will produce the same result when used with the same value of p.
In the context of TSC, Lp distances are of limited use because they cannot align two series that are misaligned in the time dimension, since they compute one-to-one differences between corresponding points only.
For example, in an electrocardiogram (ECG) signal, two measurements from a patient at different times may produce slightly different time series which belong to the same class (e.g. a certain heart condition). Ideally, if they belong to the same class, an effective similarity or distance measure should account for such “misalignments” in the time axis, while capturing the similarity or distance.
Elastic similarity and distance measures such as DTW tackle this issue. Elastic measures are designed to compensate for temporal misalignments in time series that might be due to stretched, shrunken or misaligned subsequences. From Sects. 3.2 to 3.6, we will present various elastic similarity and distance measures and show that independent and dependent strategies are substantially different.
3.2 Dynamic time warping (DTW) and related measures
3.2.1 Univariate DTW
The most widely used elastic distance measure is DTW [1]. By contrast to measures such as the Lp distance, DTW is an elastic distance measure, that allows one-to-many alignment (“warping”) of points between two time series. For many years, 1-NN with DTW was considered as the traditional benchmark algorithm for TSC [12].
DTW is efficiently solved using a dynamic programming technique. Let \(\varDelta _{DTW}\) be an \((L+1)\)-by-\((L+1)\) dynamic programming cost matrix with indices starting from \(i=0,j=0\). The first row \(i=0\) and the first column \(j=0\) defines the border conditions:
The rest of the elements \((i, j)~\textit{with}~i> 0 ~\textit{and}~j > 0\) are defined as the squared Euclidean distance between two corresponding points \(q_i\) and \(c_j\)—i.e. \(\varDelta _{DTW}(i, j) = (q_i - c_j) ^ 2\) and the minimum of the cumulative distances of the previous points. Equation (7) defines element (i, j), where \(i>0\) and \(j>0\), of the cost matrix as follows:
The cost matrix represents the alignment of the two series as according to the DTW algorithm. DTW between two series Q and C is the accumulated cost in the last element of the cost matrix (i.e.\(i,j = L +1\) ) as defined in Eq. (8):
Note that, except for LCSS, the boundary conditions of the matrix remain the same for the multivariate measures. Hence we do not repeat the boundary conditions in Eq. (6) for measures other than LCSS. We simply modify Eq. (7) accordingly to each measure.
DTW has a parameter called “window size” (w), which helps to prevent pathological warpings by constraining the maximum allowed warping distance. For example, when \(w=0\), DTW produces a one-to-one alignment, which is equivalent to the Euclidean distance. A larger warping window allows one-to-many alignments where points from one series can match points from the other series over longer time frames. Therefore, w controls the elasticity of the distance measure.
Different methods have been used to select the parameter w. In some methods, such as EE, and HIVE-COTE, w is selected using leave-one-out cross-validation. Some algorithms select the window size randomly (e.g. PF and TS-CHIEF select window sizes from the uniform distribution U(0, L/4)).
Parameter w also improves the computational efficiency, since in most cases, the ideal w is much less than the length of the series [47]. When w is small, DTW runs relatively fast, especially with lower bounding, and early abandoning techniques [48,49,50,51]. Time complexity to calculate DTW with a warping window is \(O(L \cdot w)\), instead of \(O(L^2)\) for the full DTW.
In this paper, we use DTW to refer to DTW with a cross-validated window parameter and DTWF to refer to DTW with window set to series length.
3.2.2 Dependent multivariate DTW
Definition 4
Dependent DTW (\(DTW_D\)) Dependent DTW (\(DTW_{D}\)) uses all dimensions together when computing the point-wise distance between each point in the two time series [14]. In this method, for each point in the series, DTW is allowed to warp across the dimensions.
In this case, the squared Euclidean distance between two univariate points—\((q_i - c_j) ^ 2\)—in Eq. (7) is replaced with the \(L_2\)-norm computed between the two multivariate points \(\mathbf {q_i}\) and \(\mathbf {c_j}\) as in Eq. (9).
3.2.3 Derivative DTW (DDTW)
Derivative DTW (DDTW) is a variation of DTW, which computes DTW on the first derivatives of time series. Keogh et al. [18] developed this version to mitigate some pathological warpings, particularly, cases where DTW tries to explain variability in the time series values by warping the time axis, and cases where DTW misaligns features in one series which are higher or lower than its corresponding features in the other series. The derivative transformation of a univariate time point \(q'_{i}\) is defined as:
Note that \(q'_{i}\) is not defined for the first and last element of the time series. Once the two series have been transformed, DTW is computed as in Eq. (8).
Multivariate versions of DDTW are very straightforward to implement. We calculate the derivatives separately for each dimension, and then use Eqs. (2) and (9) to compute from the derivatives independent DDTW (\(DDTW_I\)) and dependent DDTW (\(DDTW_D\)), respectively.
3.2.4 Weighted DTW (WDTW)
Weighted DTW (WDTW) is another variation of DTW, proposed by [19], which uses a “soft warping window” in contrast to the fixed warping window sized used in classic DTW. WDTW penalizes large warpings by assigning a non-linear multiplicative weight w to the warpings using the modified logistic function in Eq. (11):
where \(weight_{max}\) is the upper bound on the weight (set to 1), L is the series length, and g is the parameter that controls the penalty level for large warpings. Larger values of g increases the penalty for warping.
When creating the dynamic programming distance matrix \(\varDelta _{WDTW}\), the weight penalty \(weight_{|i{-}j|}\) for a warping distance of \(|i{-}j|\) is applied, so that the (i, j)-th ground cost in the matrix \(\varDelta _{ WDTW }\) is \(weight_{|i{-}j|}~\cdot ~(q_i~-~c_i)^2\). Therefore, the new equation for WDTW is defined as
Parameter g may be selected using leave-one-out cross-validation as in EE and HIVE-COTE, or selected randomly as in PF and TS-CHIEF (\(g\sim U(0,1)\)).
Since WDTW does not use a constrained warping window (i.e. the maximum warping distance \(|i{-}j|\) may be as large as L), its time complexity is \(O(L^2)\), which is higher than DTW.
3.2.5 Dependent multivariate WDTW
Definition 5
Dependent WDTW The dependent version of WDTW simply inserts the weight into \(DTW_D\). We define Dependent WDTW (\( WDTW _{D}\)) as
3.2.6 Weighted derivative DTW (WDDTW)
The ideas behind DDTW and WDTW may be combined to implement another measure called Weighted Derivative DTW (WDDTW). This method has also been traditionally used in some ensemble algorithms [6].
Multivariate versions of WDDTW are also straightforward to implement. We calculate the derivatives separately for each dimension, and then use Eqs. (2) and (14) with them to compute independent WDDTW (\( WDTW _I\)) and dependent WDDTW (\( WDTW _D\)), respectively.
3.3 Longest common subsequence (LCSS)
3.3.1 Univariate LCSS
Longest Common Subsequence (LCSS) distance is based on the edit distance algorithm, which is used for string matching [20, 21]. Figure 2 shows an example string matching problem. One of the early works that use LCSS for time series classification by Vlachos et al. [21] states that one motivation to use LCSS-based approach is that it is more robust to noise compared to DTW. This is because DTW matches all points, including the outliers. However, LCSS can allow some points to remain unmatched while retaining the order of matching. In addition LCSS is designed to be more efficient than DTW since it does not require the Lp computation. In TSC, the LCSS algorithm is modified to work with real-valued data by adding a threshold \(\epsilon \) for real-value comparisons. Two real values are considered a match if the difference between them is not larger than the threshold \(\epsilon \). A warping window can also be used in conjunction with the threshold to constrain the degree of local warping.
Example of string matching with LCSS. Image on the left side shows direct pairwise matching of two strings using LCSS. Image on the right side shows matching of two strings with some gaps or unmacthed letters (shown as “–”) allowed between the matched letters. This allows matching with “elasticity” as in DTW
The unnormalized LCSS distance (\(LCSS_{UN}\)) between Q and C is
In practice, \(LCSS_{UN}\) is then normalized based on the series length L.
LCSS can be used with a window parameter w similar to DTW. With a window parameter, LCSS has a time complexity of \(O(L \cdot w)\). In EE and PF, the parameter \(\epsilon \) is selected from \([\frac{\sigma }{5},\sigma ]\), where \(\sigma \) is the standard deviation of the whole dataset.
3.3.2 Independent multivariate LCSS
Independent multivariate \( LCSS_{I} \) uses the Eq. (2) and computes the LCSS for dimensions separately. However, in this case we use a separate \(\epsilon \) parameter for each dimension. We compute the standard deviation \(\sigma \) per dimension when sampling \(\epsilon \). Sampling \(\epsilon \) for each dimension can be useful if the data is not normalized.
3.3.3 Dependent multivariate LCSS
Definition 6
Dependent LCSS Dependent LCSS (\(LCSS_{D}\)) is similar to Eq. (16), except that to compute distance between two multivariate points we use Eq. (9) and an adjustment is made to the range of parameter \(\epsilon \) to make allowance for multidimensional distances tending to be larger than univariate. In this case, parameter \(\epsilon \) is selected from \([\frac{2\cdot D\cdot \sigma }{5},2\cdot D\cdot \sigma ]\), where \(\sigma \) the standard deviation of the whole dataset. Except for this adjustment, parameters are sampled similar to the way it was selected in the univariate LCSS, in EE.
Similar to the univariate case, \(LCSS_{UND}\) is then normalized based on the series length L.
3.3.4 Other \(LCSS_{D}\) formulations
An early work by Vlachos et al. [52] also presented a way to extend measures to the multivariate case. Their proposed dependent DTW is similar to Shokoohi et al.’s \({DTW_D}\) formulation in Eq. (9), but their LCSS’s formulation is slightly different to our LCSS formulation present in Eq. (19). Equation 22 defines this version of LCSS named \(Vlachos\_LCSS_{D}\).
Our method treats \(\mathbf {q_i}\) and \(\mathbf {c_i}\) as each being multidimensional points, placing a single constraint on the distance between them, whereas Vlachos et al. treat the dimensions independently, with separate constraints on each. We believe that our method is more consistent with the spirit of dependent measures.
\(Vlachos\_LCSS_{D}\) allows matching values within both data dimensions and the time dimension, while our \(LCSS_{D}\) matches values only in data dimensions. They have an additional parameter \(\delta \) that controls matching in the time dimension.
Our method also requires an adjustment to LCSS’s \(\epsilon \) parameter as described in Sect. 3.3.3. This may help to cater for unnormalized data, since \(Vlachos\_LCSS_{D}\) uses the one \(\epsilon \) across all dimensions.
3.4 Edit distance with real penalty (ERP)
3.4.1 Univariate ERP
Edit Distance with Real Penalty (ERP) [22, 23] is also based on string matching algorithms. In a typical string matching algorithm, two strings, possibly of different lengths, may be aligned by doing the least number of add, delete or change operations on the symbols. When aligning two series of symbols, the authors proposed that the delete operations in one series can be thought of as adding a special symbol to the other series. Chen et al. [22] refer to these added symbols as a “gap” element.
ERP uses the Euclidean distance between elements when there is no gap, and a constant penalty when there is a gap.
This penalty parameter for a gap is denoted as g (see Eq. (23)).
For time series, with real values, similar to the parameter \(\epsilon \) in LCSS, a floating point comparison threshold may be used to determine a match between two values. This idea was used in a measure called Edit Distance on Real sequences (EDR) [22]. However, using a threshold breaks the triangle inequality. Therefore, the same authors proposed a variant, namely ERP, which is a measure that follows the triangle inequality. Being a metric gives some advantages to ERP over DTW or LCSS in tasks such as indexing and clustering.
ERP can also be used with a window parameter w similar to DTW. With the window parameter, ERP has the same time complexity as DTW. The parameter g is selected from \([\frac{\sigma }{5},\sigma ]\), with \(\sigma \) being the standard deviation of the training data. Formally, ERP is defined as,
3.4.2 Dependent ERP
Definition 7
Dependent ERP We define Dependent ERP (\(ERP_{D}\)) as,
In Eq. (25), we note that the parameter \({\textbf{g}}\) is a vector that is sampled separately for each dimension. This is in contrast to the univariate case in Eq. (23) which uses the standard deviation of the whole training dataset (parameter g).
In this case, all terms increase proportionally with respect to the increase in the number of dimensions. So we do not need to adjust for the parameter \({\textbf{g}}\) as we adjusted for \(\epsilon \) in LCSS in Sect. 3.3.3.
3.5 Move-split-merge (MSM)
3.5.1 Univariate MSM
Move-Split-Merge (MSM) is an edit distance-based distance measure introduced by [24]. The motivation is to propose a distance measure that is a metric invariant to translations and robust to temporal misalignments. Measures such as DTW and LCSS are not metrics because they fail to satisfy the triangle inequality. Invariance to translation is another design feature of MSM when compared to ERP (see Sect. 3.4.1) [24]. For example consider two series X and Y where \(X = <v_0 \cdots v_{100}>\) and \(Y = <v>\), with v being a constant real value. ERP requires 99 deletes to transform X into Y, and the cost of deletion is tied to the value of v, since the cost of deletion is 0 if \(v=0\) or 99v otherwise (i.e. 99 merge). By contrast, in MSM, the cost of deletion is independent of the value of v. This makes MSM translation-invariant since it does not change when the same constant is added to two time series [24].
The distance between two series is computed based on the number and type of edit operations required to transform one series to the other. MSM defines three types of edit operations: move, merge and split. The move operation substitutes one value into another value. The split operation inserts a copy of the value immediately after itself, and the merge operation is used to delete a value if it directly follows an identical value. Figure 3 illustrates these edit operations.
Three edit operations used by MSM to transform one series to the other. MSM attempts to use the minimum number of these operations to perform the transformation. The number of operations used quantifies the distance between two series. Both split and merge operations incur a cost defined by the parameter c. Inspired by [24]
The cost for a move operation is the pairwise distance between two points, and the cost of split or merge operation depends on the parameter c.
Formally, MSM is defined as,
The costs of split and merge operations are defined by Eq. (29). In the univariate case, the algorithm either merges two values or splits a value if the the value of a point \(q_i\) is_between two adjacent values (\(q_{i-1}\) and \(c_j\)).
In most algorithms (e.g. EE, PF, HIVE-COTE and TS-CHIEF), the cost parameter c for MSM is sampled from 100 values generated from the exponential sequence \(\{10^{-2}, \ldots , 10^{2}\}\) proposed in Stefan et al. [24].
3.5.2 Dependent multivariate MSM
Definition 8
Dependent MSM Here we combine Eqs. (27) and (9). The cost_multiv function is explained in Sect. 3.5.3, and presented in Algorithm 1.
3.5.3 Cost function for dependent MSM
A non-trivial issue when deriving a dependent variant of MSM is how to translate the concept of one point being between two others.
A naive approach would test whether a point x is_between points y and z in multidimensional space by projecting x onto the hyperplane defined by y and z. However, this has serious limitations. For an intuitive example, let us use cities to represent points on a 2-D plane.
Assume that we have two query cities Chicago and Santiago wish to determine which is between New York and San Francisco. If we use vector projections, and project the position of Santiago on to the line between New York and San Francisco, we will find that it is between them. Similarly, we will also find that Chicago is between New York and San Francisco using this method. However, orthogonally Santiago is extremely far away from both New York and San Francisco, so it would seem more intuitive to define this function in a way that Chicago is in between New York and San Francisco, but Santiago is not. Using this intuition, we define the cost function in such a way that a point is considered to be in between two points only if the point is “inside the hypersphere” defined by the other two points. Figure 4 illustrates this concept using three points.
We implement this idea in Algorithm 1. First we find the diameter of the hypersphere in line 1 by computing \(||\mathbf {q_{i-1}} - \mathbf {c_j}||\). In line 2, we find the midpoint \(\textbf{mid}\) along the line \(\mathbf {q_{i-1}}\) and \(\mathbf {c_j}\). Then we calculate distance to the midpoint using \(||\textbf{mid} - \mathbf {q_i}||\) (line 3). Once we have the \(distance\_to\_mid\), we check if this distance is larger than half the diameter. If its larger, then the point \(\mathbf {q_i}\) is outside the hypersphere, and so we return c (line 5). If \(distance\_to\_mid\) is less than half the diameter, then \(\mathbf {q_i}\) is inside the hypersphere, so we check to which point (either \(\mathbf {q_{i-1}}\) or \(\mathbf {c_j}\)) is closest. Then we return c plus the distance to the closest point as the cost of the edit operation (line 9 to 12).
Example of checking whether a point is between two other points in 2 dimensions using a circle. In the first case (left side), the yellow point is considered “in between” blue and green points. In the second case (right side), the red point is considered to be “not in between” the green and blue points even though its projection falls between red and blue points because orthogonally it is outside the circle defined by theses points. This is one way to adapt the idea of checking if a point is between two other points in the univariate case (1-dimension) as defined in Eq. (29). In Algorithm 1, we use a generalization of this idea and check if a point is inside a hypersphere in D-dimensions (Color figure online)

3.6 Time warp edit (TWE)
3.6.1 Univariate TWE
Time Warp Edit (TWE) [25] is a further edit-distance-based algorithm adapted to the time series domain. The goal is to combine an Lp distance-based technique with an edit-distance-based algorithm that supports warping in the time axis, i.e. has some sort of elasticity like DTW, while also being a distance metric (i.e. it respects the triangle inequality). Being a metric helps in time series indexing, since it speeds up time series retrieval process.
TWE uses three operations named match, \(delete_A\), and \(delete_B\). If there is a match, Lp distance is used, and if not, a constant penalty \(\lambda \) is added. \(delete_A\) (or \(delete_B\)) is used to remove an element from the first (or second) series to match the second (or first) series. Equations 32, 33 and 34 define TWE and these three operations, respectively.
The multiplicative penalty \(\nu \)Footnote 1 is called the stiffness parameter. When \(\nu = 0\), TWE becomes more stiff like the Lp distance, and when \(\nu = \infty \), TWE becomes less stiff and more elastic like DTW. The second parameter \(\lambda \) is the cost of performing either a \(delete_A\) or \(delete_B\) operation.
Following [5, 15, 25], \(\lambda \) is selected from \(\cup _{i=0}^{9} \frac{i}{9}\) and \(\nu \) from the exponentially growing sequence \(\{10^{-5},5\cdot 10^{-5}, 10^{-4},5 \cdot 10^{-4},10^{-3},5 \cdot 10^{-3},\ldots ,1\}\), resulting in 100 possible parameterizations.
3.6.2 Dependent TWE
Definition 9
Dependent TWE Dependent version of TWE follows a similar pattern. Due to the greater magnitude of multidimensional distances, \(\lambda \) is selected from \(\cup _{i=0}^{9} \frac{2\cdot D\cdot i}{9}\) and \(\nu \) from the exponentially growing sequence \(\{2\cdot D\cdot 10^{-5},D\cdot 10^{-4},2\cdot D\cdot 10^{-4},D\cdot 10^{-3},2\cdot D\cdot 10^{-3},D\cdot 10^{-2},\ldots ,2\cdot D\}\)
We define Dependent TWE (\(TWE_{D}\)) as,
3.7 Multivariate elastic ensemble (MEE)
Ensembles formed using multiple 1-NN classifiers with a diversity of similarity and distance measures have proved to be significantly more accurate than 1-NN with any single measure [5]. Such ensembles help to reduce the variance of the model and thus help to improve the overall classification accuracy. For example, Elastic Ensemble (EE) combines eleven 1-NN algorithms, each using one of the eleven elastic measures [5]. The eleven measures used in EE are: Euclidean, DTWF (with full window), DTW (with leave-one-out cross-validated window), DDTWF, DDTW, WDTW, WDDTW, LCSS, ERP, MSM, TWE. For each measure, the parameters are optimized with respect to accuracy using leave-one-out cross-validation [5, 6]. Although EE is a relatively accurate classifier [6], it is slow to train due to the high computational cost of the leave-one-out cross-validation used to tune its parameters—\(O(n^2 \cdot L^2 \cdot P)\) for P cross-validation parameters [5, 6, 15]. Furthermore, since EE is an ensemble of 1-NN models, the classification time for each time series is also high—\(O(n \cdot L ^2)\). EE was the overall most accurate similarity or distance-based classifier on the UCR benchmark until PF [6]. EE was also used as a component of HIVE-COTE.
Next, we present our novel multivariate similarity and distance-based ensemble Multivariate Elastic Ensemble (MEE). We keep the design of our multivariate ensemble similar to the univariate EE, except that MEE uses the multivariate similarity measures. Similar to EE, MEE also uses leave-one-out cross-validation of 100 parameters when choosing the parameters for similarity and distance measures. Both EE and MEE also predict the final label of a test instance by using highest class probability. The class probability of each measure is weighted by the leave-one-out cross-validation accuracy of each measure on the training set. Any ties are broken using a uniform random choice. Similarly to the original \(DTW_I\) and \(DTW_D\) [14], all measures used in MEE use all dimensions in the dataset.
We explore four variations of MEE, which are constructed as follows.
-
\( MEE_I \): An ensemble of eleven 1-NN classifiers formed using only independent multivariate similarity and distance measures.
-
\( MEE_D \): An ensemble of eleven 1-NN classifiers formed using only dependent multivariate similarity and distance measures.
-
\( MEE_{ID} \): An ensemble of 22 1-NN classifiers formed using eleven independent measures and eleven dependent measures.
-
\( MEE_{A} \): An ensemble of eleven 1-NN classifiers formed by selecting either independent or dependent version of the measure based on its accuracy on the training set (ties are broken randomly).
4 Experiments
First we conduct experiments using 1-NN classifiers with single multivariate measures to investigate two hypotheses and then we present the experiments conducted with Multivariate Elastic Ensemble. Finally, we investigate the use of normalization and the runtime.
The first hypothesis is that there are different datasets to which each of the new multivariate distance measures is best suited. The second arises from the observation that there are datasets for which either the independent or dependent version of DTW is consistently more accurate than the alternative [14]. However, it is not clear whether this is a result of there being an advantage in treating multivariate series as either a single series of multivariate points or multiple independent series of univariate points; or rather due to some other property of the measures.
It is credible that there should be some time series data for which it is beneficial to treat multiple variables as multivariate points in a single series. Suppose, for example, that the variables each represent the throughput of independent parts of a process and the quantity relevant to classification is aggregate throughput. In this case, the sum of the values at each point is the relevant quantity. In contrast, if classification relates to a failure in any of those parts, it seems clear that independent consideration of each is the better approach.
We seek to assess whether there are multivariate datasets for which each of dependent and independent analyses is best suited, or whether there are other reasons, such as their mathematical properties, that underlie the systematic advantage on specific datasets of either \(DTW_I\) or \(DTW_D\).
We start by describing our experimental setup and the datasets we used. We then conduct an analysis of similarity and distance measures in the context of TSC by comparing accuracy measures of independent and dependent versions. We then conduct a statistical test to determine if there is a difference between independent and dependent versions of the measures.
4.1 Experimental setup
We implemented a multi-threaded version of the multivariate similarity and distance measures in Java. We also release the full source code in the github repository: https://github.com/dotnet54/multivariate-measures.
In these experiments, for parameterization of the measures, we use leave-one-out cross-validation of 100 parameters for each similarity and distance measure. We follow the same settings proposed in [5]. This parameterization is also used in HIVE-COTE, PF, and TS-CHIEF.
In this study, we use multivariate datasets obtained from https://www.timeseriesclassification.com.
For each dataset, we use 10 resamples for training with a train/test split ratio similar to the default train/test split ratio provided in the repository. Out of the available 30 datasets, we use 23 datasets in this study. Since we focus only on fixed-length datasets, the four variable length datasets (CharacterTrajectories, InsectWingbeat, JapaneseVowels, and SpokenArabicDigits) are excluded from this study. We also omit EigenWorms, MotorImagery, and FaceDetection, which take too long to run the leave-one-out cross-validation for 100 parameters in a practical time frame. Table 5 (on page 38) summarizes the characteristics of the 23 fixed-length datasets. The main results are obtained for non-normalized datasets, except for four datasets that are already normalized in the archive. We explore the influence of the normalization in Sect. 4.6. Further descriptions of each dataset can be found in [43].
4.2 Accuracy of independent vs dependent measures
First, we look at the accuracy of each measure used with a 1-NN classifier. Tables 6 and 7 (on page 39) present the accuracy for independent measures and dependent measures, respectively. For each dataset, the highest accuracy is typeset in bold. Of the values reported in Tables 6 and 7, accuracy for measures other than Euclidean distance (labeled “\(L_2\)” in the table) and DTW are newly published results in this paper.
Our first observation is that for every similarity and distance measure, except \( LCSS_D \), there is at least one dataset for which that measure obtains the highest accuracy. This is consistent with our first hypothesis, that each measure will have datasets for which it is well suited.
To compare multiple algorithms over the multiple datasets, first a Friedman test is performed to reject the null hypothesis. The null hypothesis is that there is no significant difference in the mean ranks of the multiple algorithms (at a statistical significance level \(\alpha =0.05\)). In cases where the null-hypothesis of the Friedman test is rejected, we use the Wilcoxon signed-rank test to compare the pairwise difference in ranks between algorithms, and then use Holm–Bonferroni’s method to adjust for family-wise errors [53, 54].
Figure 5 displays mean ranks (on error) between all similarity and distance measures. Measures on the right side indicate higher rank in accuracy (lower error). We do not include L2 distance here to we focus on “elastic” measures only. Since we use Holm–Bonferroni’s correction, there is not a single “critical difference value” that applies to all pairwise comparisons. Hence, we refer to these visualizations as “average accuracy ranking diagrams.” For clarity, we also remove the horizontal lines that group statistically different classifiers. We refer to the p-value tables and highlight the statistically indistinguishable pairs most relevant to the discussion in the text.
In Fig. 5, \( WDTW_D \), which is to the further right, is the most accurate measure on the evaluated datasets. \( WDTW_D \) obtained a ranking of 6.7174 from the Wilcoxon test. By contrast, \( DDTWF_D \) (ranked 14.7174) is the least accurate measure on these datasets. After Holm–Bonferroni’s correction, computed p-values indicate there are five pairs that are statistically different from each other. They are: \( DTW_D \) and \( ERP_D \), \( ERP_D \) and \( WDTW_D \), \( DDTWF_D \) and \( DTW_D \), \( DDTWF_D \) and \( WDTW_D \), \( LCSS_D \) and \( WDTW_D \).
4.3 Are independent and dependent measures significantly different?
In this section, we test if there are datasets for which independent or dependent version is always more accurate. We also test if there is a statistically significant difference between independent and dependent similarity and distance measures. Answering these questions will help us determine the usefulness of developing these two variations of the multivariate similarity and distance measures. It will also help us to construct ensembles of similarity and distance measures with more diversity, that is expected to perform well in terms of accuracy over a wide variety of datasets.
Figure 6 shows the difference in accuracy between independent and dependent versions of the measures—deeper reddish colors indicate cases where independent is more accurate (positive on the scale), and deeper bluish colors indicate cases where dependent is more accurate (negative on the scale). The datasets are sorted based on average color values to show contrasting colors on the two ends. Dimensions D, length L, number of classes c are given in the bracket after the dataset name.
Heatmap showing the difference in accuracy between independent and dependent versions of the measures—deeper reddish colors indicate cases where independent is more accurate (positive on the scale), and deeper bluish colors indicate cases where dependent is more accurate (negative on the scale). The datasets are sorted based on average color values to show contrasting colors on the two ends. (Dimensions/length/number of classes are shown in the bracket after the dataset name) (Color figure online)
From Fig. 6 we observe that there are datasets for which either independent or dependent is always more accurate. For example, the independent versions of all measures are consistently more accurate for datasets DuckDuckGeese, PEMS-SF and BasicMotions (indicated by red color rows in the heatmap). On the other hand, we see that Handwriting always wins for the dependent versions (indicated by the blue color row).
Next we statistically investigate the hypothesis that there are some multivariate TSC tasks that are inherently best suited to either treating the multivariate series as a single series of multivariate points or as multiple independent series of univariate points. To this end we present the results of a Wilcoxon signed-rank test on each of the 10 pairs of measures (without \(L_2\)), to test whether the difference between accuracy of independent and dependent versions across 23 datasets are statistically significant. We conduct this test with the null hypothesis that the mean of the difference between the accuracy of the independent and dependent versions will be zero. We reject the null hypothesis with statistical significance value \(\alpha =0.05\), and accept that there is a significant statistical difference in accuracy if the \(p\le \alpha \). Table 2 shows the p-value for each dataset. The bold values mark the p-values for which there is a significant difference. We also report the adjusted \(\alpha \) value after Holm–Bonferroni corrections, \(\alpha _{HB}\). Before Holm–Bonferroni correction, out of the 23 datasets, we observe that for 8 datasets there is a statistically significant difference in accuracy between independent and dependent measures. After Holm–Bonferroni correction, we still find 5 statistically significant differences (where \(p\le \alpha _{HB}\)) and so conclude there are indeed datasets that are inherently best suited to either independent or dependent treatment.
This finding leads to the questions of why this is the case and whether it is predictable which strategy will prevail.
In general, we observe that the dependent strategy tends to perform poorly when there are a large number of dimensions in the dataset. The two datasets for which the independent strategy performs most strongly are the only two with more than 100 dimensions. The explanation for this may simply be that high-dimensional datasets suffer from the curse of dimensionality and that the \(L_2\)-norms between different sets of high dimensional points carry little information.
For lower-dimensional data, we hypothesize that the independent strategy will be effective when the interactions between the dimensions carry little information about the classification task and the dependent strategy will be effective when the interactions between the dimensions carry much information about the classification task.
Consider the Handwriting dataset for which all dependent measures are more accurate than their corresponding independent measures (see Fig. 6). It has three accelerometer values recording hand written letters. It seems intuitive that to distinguish a straight line from curved; horizontal from vertical; and writing from repositioning the pen; it is necessary to consider all three accelerometers together. To test this, we compare the accuracy of 1-NN classifiers using each dimension alone to the accuracy of the independent and dependent strategies. The results are shown in Table 3. Firstly, we observe that for Handwriting dataset, \(DTW_D\) performs more accurately than \(DTW_I\) (0.61 versus 0.46). In addition, we see that using \(DTW_D\) with all three dimensions of the dataset is more accurate than using only a single dimension (last three rows). This shows that dependent measures can be effective for datasets that contain information in the interactions between dimensions.
We contrast this to the six dimensional BasicMotions dataset for which all independent measures are more accurate than their corresponding dependent measures (see Fig. 6). Comparing the accuracy of the single-dimension 1-NN classifiers to that of the independent and dependent strategies, we see that each of three of the dimensions when used alone can attain 100% accuracy. When all dimensions are considered together, \(DTW_I\) also performs better than \(DTW_D\) (1.00 versus 0.96). This suggests that independent measures will have good performance when individual dimensions carry substantial information about the class without need to consider interdependencies between dimensions.
It is interesting to note that there appears to be considerable correlation between the relative desirability of the independent and dependent approaches across all the measures that are applied to the derivative of the original series, DDTWF, DDTW and WDDTW. It particularly stands out that there seems to be a strong advantage to independent variants of these measures with respect to ArticularyWordRecognition, DuckDuckGeese and Cricket. Possible connections between transformations and the relative efficacy of independent or dependent approaches may be a productive topic for future research.
4.4 Multivariate measures vs MEE
EE showed that 1-NN ensembles formed using a diverse set of similarity and distance measures are more accurate than any of the single measures on the 85 univariate UCR datasets [5, 6]. In this experiment, we replicate this evaluation in the multivariate context. We compare single independent and dependent measures with MEE ensembles to assess whether each of the three versions of MEE are significantly different to the individual measures.
Figures 7 and 8 show accuracy rankings of independent measures and \( MEE_I \); and dependent measures and \( MEE_D \), respectively. Figure 7 indicates that \( MEE_I \) obtains the best accuracy. However, an investigation of the p-values indicate that \( MEE_I \) is not significantly different from seven independent measures (\( WDTW_I \), \( DTW_I \), \( DTWF_I \), \( MSM_I \), \( ERP_I \), \( WDDTW_I \), and \( DDTWF_I \)). As for, Fig. 8, the computed p-values indicate that \( MEE_D \) is significantly different to individual measures except for \( WDTW_D \),\(DTWF_D\) and \(DTW_D\). Both results indicate that \( MEE_I \) and \( MEE_D \) are more accurate than individual measures with 1-NN.
Figure 9 shows accuracy ranking of our four ensembles presented in Sect. 3.7versus the top five (out of twenty-one) individual similarity and distance measures with 1-NN. We can observe that all ensembles are more accurate than the classifiers using a single measure. We found that the \( MEE_{A} \) (avg. rank 2.5870) performs better than \( MEE_{ID} \) (avg. rank 2.9130), \( MEE_I \) (avg. rank 3.7174) and \( MEE_D \) (avg. rank 5.0435). The computed p-values show that the difference between \( MEE_{A} \) and \( MEE_D \) is statistically significant. Based on these results, we select \( MEE_{A} \) as the final design of \( MEE \).
4.5 MEE vs SOTA multivariate TSC algorithms
Next, we compare \( MEE \) (i.e. \( MEE_A \)) with six state-of-the-art multivariate TSC algorithms [29, 42]. Figure 10 shows the accuracy ranking of our ensembles and these algorithms.
The most accurate algorithm, HIVE-COTE 2.0, obtained an average rank of 2.7826. It is followed by ROCKET (ranked 3.3696), InceptionTime (3.6957), DrCIF (ranked 3.7391) and then our ensemble \( MEE \) (4.5000). Our ensemble \( MEE \) is not significantly different to more complex leading classifiers such as HIVE-COTE 2.0, ROCKET and InceptionTime. Based on the p-values, only 2 pairs of classifiers are significantly different. They are: STC vs HIVE-COTE 2.0 and TDE vs HIVE-COTE 2.0.
Finally, Table 4 shows the accuracy of leading algorithms (selected from Fig. 10) and our ensembles. Across the all algorithms, \( MEE \) achieves the highest accuracy for 2 datasets. By comparison, shapelet-based STC is highest on 1 dataset and dictionary-based TDE on 2 datasets. Interestingly, highly ranked ROCKET has highest accuracy on only two datasets, while InceptionTime and HIVE-COTE 2.0 have highest accuracy six and eight times, respectively.
4.6 Performance on normalized vs unnormalized data
We ran experiments for all measures for both z-normalized and unnormalized datasets. Note that 4 datasets—ArticularyWordRecognition, Cricket, HandMovementDirection, and UWaveGestureLibrary—are already normalized. These were excluded from this study as it is not possible to derive unnormalized versions. We z-normalized each of the remaining datasets on a per series, per dimension basis. We found that the accuracy is higher without normalization. This agrees with a recent paper which conducted a similar experiment using \(DTW_I\) and \(DTW_D\) [42].
We include the results for all measures in our github repository.Footnote 2 To summarize the results, Fig. 11 shows a scatter plot comparing the accuracy of MEE with normalized and unnormalized data. This comparison was conducted on the default train and test split and excludes the already normalized four datasets in the archive. The results show MEE with unnormalized datasets win 14 times and loses 4 times with 1 tie. In addition, for both \( MEE_I \) (12/2/5 win/draw/loss) and \( MEE_D \) (11/1/7 win/draw/loss) we also observed that unnormalized data works better. This is why we used unnormalized datasets for all other experiments in the paper.
However, we note that this does not indicate that not normalizing is always the optimal solution for all datasets. Sometimes normalization can be useful when using similarity and distance measures. For example, consider a scenario with two dimensions temperature (e.g a scale from 0 to 100 degree Celsius) and relative humidity as a proportion (between 0 and 1). In such a case, temperature will dominate the result of the similarity or distance calculation, and normalization will help to compute the similarity or distance with similar scales across the dimensions.
4.7 Runtime
In this section, we provide a summary of the runtime information and discuss the time complexity of the multivariate measures and the MEE.
We ran the experiments on a cluster of Intel(R) Xeon(R) CPU E5-2680 v3 @ 2.50 GHz CPUs, using 16-threads. The total time to train 23 datasets with leave-one-out cross-validation with 10 resamples of the training set was about 10,423 h (wall clock time). On average, one fold of leave-one-out cross-validation required 1042 h of train time. The two slowest datasets were PEMS-SF (149 h) and PhonemeSpectra (650 h) per fold, about 76% of the total training time. Moreover, 11 out of 23 datasets took less than one hour per dataset to train.
The slowest measure to train was \(MSM_D\), which took a total of 3553 h across all datasets and the 10 folds (almost 35% of the total training time). The second slowest measure \(MSM_I\) took 1109 h, and the third slowest \(TWE_I\) took 851 h. By contrast, the fastest measure to train was \(DTWF_D\) and took just one hour. This is excluding Euclidean distance with no parameter which only took few minutes. Note that since MEE uses training accuracy to weight the ensemble predictions, leave-on-out cross-validation is performed at least once even if there are no parameters or a single parameter (e.g. DTWF). Figure 12 shows the average runtime of each measure per fold.
Compared to classifiers such as ROCKET, our ensemble, MEE created from these measures have very high training time. However, the goal of MEE is not to tackle the scalability issue but to create a multivariate similarity and distance-based classifier as a baseline for accuracy comparison with other similarity and distance-based multivariate classifiers. This is similar to the univariate EE, which was extremely slow with a time complexity of \(O(n^2 \cdot L^2 \cdot P)\) for P cross-validation parameters [5, 6, 15]. Despite its speed, EE stimulated much research in TSC and helped the development of more accurate classifiers such as the earlier versions of HIVE-COTE, and much faster classifiers such as PF and TS-CHIEF. Training time and test time complexity of MEE is: \(O(n^2 \cdot L^2 \cdot D \cdot P)\) and \(O(n \cdot L^2 \cdot D)\), respectively.
Measures such as DTW can be scaled to millions of time series when used in conjunction with lower bounding and early abandoning [12, 50, 55]. Therefore, if various research on lower bounding and early abandoning of other similarity and distance measures [48, 49, 51, 56] are combined and extended to multivariate measures, then it might be possible to create a more scalable version of MEE.
5 Conclusion
In this paper, we present multivariate versions of seven commonly used elastic similarity and distance measures. Our approach is inspired by independent and dependent DTW measures, which have proved very successful as strategies for extending univariate DTW to the multivariate case.
These measures can be used in a wide range of time series analysis tasks including classification, clustering, anomaly detection, indexing, subsequence search and segmentation. This study demonstrates their utility for time series classification. Our experiments show that each of the univariate similarity and distance measures excels at nearest neighbor classification on different datasets, highlighting the importance of having a range of such measures in our analytic toolkits.
They also show that there are datasets for which the independent version of DTW is more accurate than the dependent version and vice versa. Until now there was no way to determine whether this is a result of a fundamental difference between treating each dimension independently or not, or whether it arises from other properties of the algorithms. Our results showing that there are some datasets for which dependent or independent treatments are consistently superior across all distance measures provides strong support for the conclusion that it is a fundamental property of the datasets, that either the variables are best considered as a single multivariate point at each time step or are not.
We observe that the dependent strategy tends to perform poorly when there are a large number of dimensions in the dataset. Addressing this limitation may be a productive direction for future research. We further observe that the independent method tends to perform well when individual dimensions are independently accurate univariate classifiers and that it is credible that dependent approaches excel when there are strong mutual interdependencies between them with respect to the class.
Inspired by the Elastic Ensemble of nearest neighbor classifiers using different univariate distance measures, we then further experiment with ensembles of multivariate similarity and distance measures and show that ensembling results in accuracy competitive with the state of the art.
Our three ensembles establish a baseline in our future plans to create a multivariate TS-CHIEF which would combine similarity and distance-based techniques with dictionary-based, interval-based for multivariate TSC.
Notes
In the published definition of TWE [25], \(\nu \) is multiplied with the time difference in the timestamps of two consecutive time points. We simplified this equation, for clarity, by assuming that this time difference is always 1 (UEA datasets do not contain the actual timestamps).
References
Sakoe H, Chiba S (1978) Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans Acoust Speech Signal Process 26(1):43–49
Berndt DJ, Clifford J (1994) Using dynamic time warping to find patterns in time series. In: Proceedings of AAAI workshop on knowledge discovery in databases, vol 10. Seattle, WA, USA, pp 359–370
Aghabozorgi S, Shirkhorshidi AS, Wah TY (2015) Time-series clustering-a decade review. Inf Syst 53:16–38
Liao TW (2005) Clustering of time series data-a survey. Pattern Recognit 38(11):1857–1874
Lines J, Bagnall A (2015) Time series classification with ensembles of elastic distance measures. Data Min Knowl Disc 29(3):565–592
Bagnall A, Lines J, Bostrom A, Large J, Keogh E (2017) The great time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data Min Knowl Disc 31(3):606–660
Izakian H, Pedrycz W (2014) Anomaly detection and characterization in spatial time series data: a cluster-centric approach. IEEE Trans Fuzzy Syst 22(6):1612–1624
Steiger M, Bernard J, Mittelstädt S, Lücke-Tieke H, Keim D, May T, Kohlhammer J (2014) Visual analysis of time-series similarities for anomaly detection in sensor networks. In: Computer graphics forum, vol 33. Wiley Online Library, pp 401–410
Gunopulos D, Das G (2001) Time series similarity measures and time series indexing. ACM SIGMOD Rec 30(2):624
Park S, Kim S-W, Chu WW (2001) Segment-based approach for subsequence searches in sequence databases. In: Proceedings of the 2001 ACM symposium on Applied computing, pp 248–252
Cassisi C, Montalto P, Aliotta M, Cannata A, Pulvirenti A (2012) Similarity measures and dimensionality reduction techniques for time series data mining. Advances in Data Mining Knowledge Discovery and Applications (InTech Rijeka, Croatia 2012), 71–96
Ding H, Trajcevski G, Scheuermann P, Wang X, Keogh E (2008) Querying and mining of time series data: experimental comparison of representations and distance measures. Proc VLDB Endow 1(2):1542–1552
Keogh E, Kasetty S (2003) On the need for time series data mining benchmarks: a survey and empirical demonstration. Data Min Knowl Disc 7(4):349–371
Shokoohi-Yekta M, Hu B, Jin H, Wang J, Keogh E (2017) Generalizing DTW to the multi-dimensional case requires an adaptive approach. Data Min Knowl Disc 31(1):1–31
Lucas B, Shifaz A, Pelletier C, O’Neill L, Zaidi N, Goethals B, Petitjean F, Webb GI (2019) Proximity forest: an effective and scalable distance-based classifier for time series. Data Min Knowl Disc 33(3):607–635
Lines J, Taylor S, Bagnall A (2018) Time series classification with HIVE-COTE: the Hierarchical vote collective of transformation-based ensembles. ACM Trans Knowl Discovery Data, 12(5)
Shifaz A, Pelletier C, Petitjean F, Webb GI (2020) TS-CHIEF: a scalable and accurate forest algorithm for time series classification. Data Min Knowl Disc 34(3):742–775
Keogh EJ, Pazzani MJ (2001) Derivative dynamic time warping. In: Proceedings of the 2001 SIAM international conference on data mining. SIAM, pp 1–11
Jeong Y-S, Jeong MK, Omitaomu OA (2011) Weighted dynamic time warping for time series classification. Pattern Recognit 44(9):2231–2240
Hirschberg DS (1977) Algorithms for the longest common subsequence problem. J ACM 24(4):664–675
Vlachos M, Kollios G, Gunopulos D (2002) Discovering similar multidimensional trajectories. In: Proceedings 18th international conference on data engineering. IEEE, pp 673–684
Chen L, Ng R (2004) On the marriage of lp-norms and edit distance. In: Proceedings of the thirtieth international conference on VLDB-volume 30, pp 792–803
Chen L, Özsu MT, Oria V (2005) Robust and fast similarity search for moving object trajectories. In: Proceedings of the 2005 ACM SIGMOD international conference on management of data, pp 491–502
Stefan A, Athitsos V, Das G (2012) The move-split-merge metric for time series. IEEE Trans Knowl Data Eng 25(6):1425–1438
Marteau P-F (2008) Time warp edit distance with stiffness adjustment for time series matching. IEEE Trans Pattern Anal Mach Intell 31(2):306–318
Itakura F (1975) Minimum prediction residual principle applied to speech recognition. IEEE Trans Acoust Speech Signal Process 23(1):67–72
Deng H, Runger G, Tuv E, Vladimir M (2013) A time series forest for classification and feature extraction. Inf Sci 239:142–153
Middlehurst M, Large J, Bagnall A (2020) The canonical interval forest (CIF) classifier for time series classification. In: 2020 IEEE international conference on big data (big data). IEEE, pp 188–195
Middlehurst M, Large J, Flynn M, Lines J, Bostrom A, Bagnall A (2021) HIVE-COTE 2.0: a new meta ensemble for time series classification. Mach Learn 110(11):3211–3243
Hills J, Lines J, Baranauskas E, Mapp J, Bagnall A (2014) Classification of time series by shapelet transformation. Data Min Knowl Disc 28(4):851–881
Karlsson I, Papapetrou P, Boström H (2016) Generalized random shapelet forests. Data Min Knowl Disc 30(5):1053–1085
Bagnall A, Flynn M, Large J, Lines J, Middlehurst M (2020) On the usage and performance of the Hierarchical Vote Collective of Transformation-based Ensembles version 1.0 (HIVE-COTE v1. 0). In: International workshop on advanced analytics and learning on temporal data. Springer, Berlin, pp 3–18
Lin J, Khade R, Li Y (2012) Rotation-invariant similarity in time series using bag-of-patterns representation. J Intell Inf Syst 39(2):287–315
Schäfer P (2015) The BOSS is concerned with time series classification in the presence of noise. Data Min Knowl Disc 29(6):1505–1530
Schäfer P, Leser U (2017) Fast and accurate time series classification with weasel. In: Proceedings of the 2017 ACM on conference on information and knowledge management, pp 637–646
Middlehurst M, Large J, Cawley G, Bagnall A (2020) The temporal dictionary ensemble (TDE) classifier for time series classification. In: Joint European conference on machine learning and knowledge discovery in databases. Springer, Berlin, pp 660–676
Dempster A, Petitjean F, Webb GI (2020) ROCKET: exceptionally fast and accurate time series classification using random convolutional kernels. Data Min Knowl Disc 34(5):1454–1495
Gallicchio C, Micheli A (2017) Deep echo state network (deepesn): a brief survey. arXiv preprintarXiv:1712.04323
Wang Z, Yan W, Oates T (2017) Time series classification from scratch with deep neural networks: a strong baseline. In: International joint conference on neural networks (IJCNN). IEEE 2017:1578–1585
Fawaz HI, Lucas B, Forestier G, Pelletier C, Schmidt DF, Weber J, Webb GI, Idoumghar L, Muller P-A, Petitjean F (2020) Inceptiontime: finding AlexNet for time series classification. Data Min Knowl Disc 34(6):1936–1962
Fawaz HI, Forestier G, Weber J, Idoumghar L, Muller P-A (2019) Deep learning for time series classification: a review. Data Min Knowl Disc 33(4):917–963
Ruiz AP, Flynn M, Large J, Middlehurst M, Bagnall A (2021) The great multivariate time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data Min Knowl Disc 35(2):401–449
Bagnall A, Dau HA, Lines J, Flynn M, Large J, Bostrom A, Southam P, Keogh E (2018) The UEA multivariate time series classification archive, 2018. arXiv preprintarXiv:1811.00075
Middlehurst M, Vickers W, Bagnall A (2019) Scalable dictionary classifiers for time series classification. In: International conference on intelligent data engineering and automated learning. Springer, pp 11–19
Löning M, Bagnall A, Ganesh S, Kazakov V, Lines J, Király FJ (2019) Sktime: a unified interface for machine learning with time series. In: Workshop on systems for ML at NeurIPS 2019
Zhang X, Gao Y, Lin J, Lu C-T (2020) Tapnet: multivariate time series classification with attentional prototypical network. Proc AAAI Conf Artif Intell 34(04):6845–6852
Tan CW, Herrmann M, Forestier G, Webb GI, Petitjean F (2018) Efficient search of the best warping window for dynamic time warping. In: Proceedings of the 2018 SIAM international conference on data mining. SIAM, pp 225–233
Keogh E, Wei L, Xi X, Vlachos M, Lee S-H, Protopapas P (2009) Supporting exact indexing of arbitrarily rotated shapes and periodic time series under Euclidean and warping distance measures. VLDB J 18(3):611–630
Lemire D (2009) Faster retrieval with a two-pass dynamic-time-warping lower bound. Pattern Recognit 42(9):2169–2180
Tan CW, Webb GI, Petitjean F (2017) Indexing and classifying gigabytes of time series under time warping. In: Proceedings of the 2017 SIAM international conference on data mining. SIAM, pp 282–290
Herrmann M, Webb GI (2021) Early abandoning and pruning for elastic distances including dynamic time warping. Data Min Knowl Disc 35(6):2577–2601
Vlachos M, Hadjieleftheriou M, Gunopulos D, Keogh E (2003) Indexing multi-dimensional time-series with support for multiple distance measures. In: Proceedings of the ninth ACM SIGKDD international conference on knowledge discovery and data mining, pp 216–225
Demšar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Benavoli A, Corani G, Mangili F (2016) Should we really use post-hoc tests based on mean-ranks? J Mach Learn Res 17(1):152–161
Keogh E, Wei L, Xi X, Lee S-H, Vlachos M (2006) LB_Keogh supports exact indexing of shapes under rotation invariance with arbitrary representations and distance measures. In: Proceedings of the 32nd international conference on very large databases. Citeseer, pp 882–893
Tan CW, Petitjean F, Webb GI (2019) Elastic bands across the path: a new framework and method to lower bound dtw. In: Proceedings of the 2019 SIAM international conference on data mining. SIAM, pp 522–530
Acknowledgements
This research was supported by the Australian Research Council under grant DP210100072. This material is based upon work supported by the Air Force Office of Scientific Research, Asian Office of Aerospace Research and Development (AOARD) under award number FA2386-18-1-4030.
The authors would like to thank Prof. Anthony Bagnall and everyone who contributed to the UEA multivariate time series classification archive.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research has been supported by Australian Research Council grant DP210100072.
Appendices
Summary of the Datasets
See Tables 5.
Accuracy of dependent and independent measures
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shifaz, A., Pelletier, C., Petitjean, F. et al. Elastic similarity and distance measures for multivariate time series. Knowl Inf Syst 65, 2665–2698 (2023). https://doi.org/10.1007/s10115-023-01835-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-023-01835-4