Abstract
This study aimed to identify the differences presented in the Raman spectrum of blood serum from normal subjects compared to leukemic and non-leukemic subjects and the differences between the leukemics and non-leukemics, correlating the spectral differences with the biomolecules. Serum samples from children and adolescents were subjected to Raman spectroscopy (830 nm, laser power 350 mW; n = 566 spectra, being 72 controls, 269 leukemics, and 225 non-leukemics). Exploratory analysis based on principal component analysis (PCA) of the serum sample’s spectra was performed. Classification models based on partial least squares discriminant analysis (PLS-DA) were developed to classify the spectra into normal, leukemic, and non-leukemic, as well as to discriminate spectra of leukemic from non-leukemic. The exploratory analysis showed principal components with peaks related to amino acids, proteins, lipids, and carotenoids. The spectral differences between normal, leukemic, and non-leukemic showed features assigned to proteins (serum features), amino acids, and carotenoids. The PLS-DA model classified the spectra of the normal group versus leukemic and non-leukemic groups with accuracy of 66%, sensitivity of 99%, and specificity of 57%. The PLS-DA discriminated the spectra of the leukemic and non-leukemic groups with accuracy of 67%, sensitivity of 72%, and specificity of 60%. The study showed that Raman spectroscopy is a technique that may be used for the biochemical differentiation of leukemias and other types of cancer in serum samples of children and adolescents. Nevertheless, building an extensive data library of Raman spectra from serum samples of controls, leukemics, and non-leukemics of different age groups is necessary to understand the findings better.
Similar content being viewed by others
Introduction
Cancer arises from a disordered multiplication of altered cells that form a tumor and is constituted by a set of heterogeneous genetic instabilities united by common alterations in multiple cell signaling pathways [1, 2]. Cancer is a major public health problem in the world, like hypertension and diabetes [3], and is already among the four leading causes of premature death (before 70 years of age) in most countries [2, 4]. Cancer is the second most common cause of death among children aged 1 to 14 years in the United States. In 2021, the American Cancer Society estimated that 10,500 children (aged birth to 14 years) and 5,090 adolescents (aged 15–19 years) will be diagnosed with cancer [4]. According to the Brazilian National Cancer Institute, the prevision of cancer incidence in Brazil for the triennium 2020–2022 indicates the occurrence of approximately 625,000 new cases of cancer [2]. Cancer types and their distribution in adolescents differ from those in children; there are almost twice as many cases of Hodgkin as non-Hodgkin lymphoma among adolescents, whereas it is the reverse among children. Leukemia is the most common childhood cancer, accounting for 28% of cases, followed by brain and other nervous system tumors (27%) [4].
Cancer in children and adolescents is difficult to detect and presents various signs or symptoms, many of which are confused with common childhood illnesses [5, 6]. Cancer represents an important barrier to increasing life expectancy in the world [4]. The impact of the coronavirus disease 2019 (COVID-19) pandemic, as well as its extent in different parts of the world, is still unknown, but it is assumed that delays in diagnosis and treatment, associated with the concerns of individuals and the closure of systems health care, with reduced care assistance, cause a short-term decline in cancer incidence, followed by increases in late-stage diagnoses and cancer mortality in some settings [4].
Childhood cancers are rare, and there are no widely recommended screening tests to diagnose cancer in children. Early diagnosis is usually based on symptoms, clinical history, laboratory tests, and imaging (radiographs, tomography, and magnetic resonance). Confirmation is made by excisional biopsy following anatomopathology [2, 4, 7]. In the case of leukemias and lymphomas, a bone marrow aspirate is collected in addition to tissue histopathological analysis (biopsy). Bone marrow aspirate allows detailed cell morphology examination and blast percentage assessment. Bone marrow biopsy allows bone marrow cellularity and architecture to be determined [4, 8,9,10].
In recent years, optical spectroscopy techniques such as fluorescence, elastic scattering, and Raman spectroscopy were proposed for rapid and non-invasive medical diagnosis [10,11,12,13,14,15,16,17]. Such techniques can provide information on the metabolites present in tissues (fluorescence of blood plasma aiming detection of porphyrins for diagnosis of colorectal cancer) [15], detecting blood (hemoglobin) content in tissues (elastic scattering of sentinel lymph nodes for breast cancer diagnosis) [16] and molecular composition of biological tissues and fluids (Raman spectroscopy of skin cancer and its biochemicals aiming diagnosis) [17], with Raman spectroscopy being a feasible technique for providing a detailed biochemical composition of cells, tissues, and fluids even in vivo aiming at diagnosis [10,11,12,13,14, 17, 18].
Raman spectroscopy is based on the inelastic scattering of incident laser light by the molecules, and studies demonstrated the possibilities of the technique to determine the biochemical composition of biological tissues and fluids for qualitative (differentiation in the biochemical composition) and quantitative (differences in the concentration of selected biochemicals) analyses [10, 11, 17, 19, 20]. It can be used for cancer diagnosis [17, 19, 21,22,23,24,25] and cancer prognosis [13, 26, 27], being considered promising for showing the differences and using them to discriminate between benign and malignant tissues in different pathologies, including prostate [21, 22], stomach [23, 24], breast cancer [25], and skin [14, 19, 26], among others [11, 13, 17, 20, 27] in both in vivo and ex vivo. Raman technique does not require complicated sample preparation; it is non-invasive and non-destructive and provides information on the vibrational energy modes of molecules in real-time, allowing in situ analysis of biological tissues with high precision [18, 20, 21] without tissue removal [11, 12, 17, 19, 28, 29]. Near-infrared excitation (between 785 and 830 nm) for Raman spectroscopy studies in biological tissues has the advantage of low fluorescence emission due to the low absorption [11, 30].
Studies with Raman spectroscopy for blood-related investigations began in the 1970s when Goheent et al. [31] compared Raman spectra (514.5 nm excitation) of intact erythrocyte membranes with spectra of membranes from which essentially, all peripheral proteins were extracted. Atkins et al. [32] used Raman spectroscopy for the biochemical characterization of blood and its components. In a study with blood serum samples, González-Solís et al. [20] used Raman and principal component analysis (PCA) to differentiate serum from normal and leukemic patients and identify the different types of leukemia based on the biochemical composition of the serum. Silva et al. [27] used whole blood and plasma samples from healthy and leukemic subjects based on Raman and PCA to differentiate leukemic and non-leukemic patients according to biochemical changes. González-Solís et al. [20] and Silva et al. [27] showed an increase in the concentration of plasma carotenoids in the normal group and associated this compound with a protective role against neoplasms. Martínez-Espinosa et al. [33] collected Raman spectra from serum samples from leukemia patients and healthy volunteers to discriminate them and used PCA in the Raman spectra to be able to differentiate the samples. Bai et al. [34] used Raman spectroscopy to analyze the characteristics of blood plasma from patients with diffuse large B-cell lymphoma (DLBCL) and chronic lymphocytic leukemia (CLL) compared to plasma from healthy patients. Using orthogonal partial least squares discriminant analysis (OPLS-DA), they obtained good discrimination among the DLBCL, CLL, and healthy volunteers (control) groups; the authors also found Raman bands specific for patients with DLBCL and CLL. Therefore, the Raman spectral characteristics of the blood serum combined with the multivariate discriminant analysis of spectral data using PCA can show the differences between groups (normal and leukemic, for instance) and can be used in the diagnosis through Raman spectroscopy.
This study aimed to use Raman spectroscopy (830 nm excitation) to identify spectral differences in the blood serum of healthy (normal group), leukemic (leukemic group), and non-leukemic (non-leukemic group) children and adolescents. The spectral differences related to the biochemical components presented in each group were statistically evaluated by the ANOVA test, and then these peaks were assigned to their corresponding chemical compositions already described in the scientific literature. The spectral dataset was subjected to exploratory analysis based on PCA, and then a classification model was developed employing partial least squares discriminant analysis (PLS-DA) for the spectral differentiation of normal, leukemic, and non-leukemic samples. The present study highlights the differentiation of Raman spectral data obtained in blood serum samples from healthy children and adolescents and from ones with leukemia and with other oncological pathologies.
Materials and methods
Ethics committee
The protocol used in this study was approved by the Research Ethics Committee of University Anhembi Morumbi—UAM (Process CAAE No. 23457019.9.0000.5492) following the Brazilian guidelines for research in humans. All patients submitted to the research signed a Free and Informed Consent Term (FICT), which informed the entire procedure to be performed and the risks and benefits of the research.
Patient selection
Blood serum samples from children and adolescents were selected from the Group of Attention to Children with Cancer (GACC) clinical analysis laboratory in São José dos Campos, SP. The study included children and adolescents between 0 and 19 years of age (9.8 ± 6.2 years of age) among the patients who underwent surgical treatment for oncological and non-oncological pathologies treated at GACC and patients who were submitted to elective surgeries. Patients with oncological pathologies were diagnosed with acute leukemias or other non-leukemic oncological pathologies (sarcomas, blastomas, carcinomas, and lymphomas) in different stages of treatment. The patients submitted to elective surgeries were considered healthy from a hematological point of view. Patients who refused to participate in the research, patients who did not have blood material in adequate conditions for analysis, and patients in the normal group who had associated infectious diseases were excluded from the study.
Blood samples were collected from peripheral veins of subjects by a vacuum-closed method in tubes without anticoagulant and with clot activator-containing gel (model Vacuette®, Greiner Bio-One Brasil Ltda., SP, Brazil). Five mL of peripheral blood was collected and then centrifuged at 3.000 rpm for 15 min to separate the serum from the cells. The tubes with centrifuged samples were stored in a freezer (temperature from − 25 to − 18 °C) to minimize alterations in the biochemical constitution of serum.
Serum samples from the patients were divided into three groups: normal group (healthy patients from elective surgeries without an established infectious or inflammatory process), leukemic group (patients diagnosed with acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML)), and non-leukemic group (patients diagnosed with sarcoma, blastoma, carcinoma, and lymphoma), according to the anatomopathological diagnoses extracted from medical records. A total of 186 samples of serum were evaluated: 21 samples from the normal group, 90 samples from the leukemic group, and 75 samples from the non-leukemic group (Table 1). On the day of acquisition of the Raman spectra, samples were thawed at room temperature, and analyses were performed in 80 µL of serum.
Raman spectrometer and data acquisition
The Raman spectrometer used in the study (model Dimension P1, Lambda Solutions Inc., MA, USA) has excitation wavelength of 830 nm, power adjustable up to 450 mW, and working temperature of − 75 °C (thermoelectric cooling). The spectrometer has a diffraction grating of 1,200 lines/mm that promotes the dispersion of scattered light onto the back-thinned, deep-depletion CCD (charge-coupled device) detector (1,340 × 100 pixels) with a resolution of 2.5 cm−1 in the spectral range between 400 and 1800 cm−1. The spectrometer uses a Raman probe (model Vector Probe, Lambda Solutions Inc.) for sample excitation and signal acquisition, allowing repeatable excitation-collection geometry. The use of near-infrared excitation (830 nm) minimizes the emission (fluorescence) due to the presence of absorbers in biological samples. The power used in the probe’s output was adjusted to 350 mW.
The Raman shift calibration was verified at the beginning of the experiment by taking the naphthalene spectrum. The spectrometer’s spectral response and probe were corrected using an intensity calibrated tungsten lamp (a description can be found in Silveira et al. [13]).
Raman spectra were obtained from blood serum samples without any preparation. 80 μL of the serum stored in the test tube was pipetted in a clean aluminum sample holder with holes using a disposable tip (model P200, Bio-Rad Laboratorios Brasil Ltda., SP, Brazil). The exposure time for obtaining each spectrum was 3 s with 10 accumulations (30-s total exposure time). The spectrum of each sample was taken in three (samples from the normal, leukemic, and non-leukemic groups) to six (samples from the normal group) replicates for statistical purposes by moving the sample holder laterally at different points.
The collected Raman spectra were subjected to pre-processing to remove the Raman background (mainly fluorescence) by fitting and subtracting a sixth-order polynomial over the entire spectral range of 400 to 1800 cm−1, to each spectrum in a routine implemented in MATLAB 7.01 software (version 2007a, The MathWorks Inc., MA, USA). Cosmic rays spikes were removed manually, and then the spectra were normalized by the area under the curve (1-norm) [35]. The pre-processing is an important step to reduce interferences that may occur during the spectra acquisition, such as cosmic rays, background fluorescence, and eventual changes in laser power. After processing, one spectrum from the leukemic group was excluded due to low signal-to-noise ratio. The final dataset corresponded to 566 Raman spectra from 186 serum samples (Table 1).
The most intense peaks in the mean Raman spectra of normal, leukemic, and non-leukemic groups were marked and attributed to the biochemical elements of the blood serum, according to the published literature [20, 27, 33, 34, 36,37,38] (peaks described in Table 2). Statistical analysis was performed to assess which peaks presented significant differences between the groups. First, Levine’s test checked the Gaussian (normal) distribution. Then, the one-way analysis of variance (ANOVA) with Tukey–Kramer post-test or Kruskal–Wallis’s test with Dunn’s test was applied to the peaks with Gaussian distribution and non-Gaussian distribution, respectively. Differences were considered significant when p value < 0.05.
Exploratory analysis by PCA
PCA is a statistical method that reduces the data dimensionality and preserves as much variability as possible within the dataset; it is ideal for working with large, multivariate data. PCA finds new variables based on the variance of the dataset, being linear functions of the original data that successively maximize the variance (one variable is not correlated with the other). The new variables depend on the dataset rather than being predefined functions and therefore are adaptive in the broad sense [39, 40]. The first principal component corresponds to the direction along which the samples show the highest spectral variance; the second principal component is the direction uncorrelated with the first component along which the samples point to the second highest variance [39, 40] and so on. PCA generates two new variables: principal component scores (SCs), which resemble a Raman spectrum, and loadings (PCs), which indicate the intensity of each score present in the original data. From the analysis of these two variables, the differences and similarities between the studied groups can be identified [27, 35, 39, 40]. In this study, the exploratory analysis was performed with the PCA to identify which scores (score 1 to score 8) presented spectral differences between the normal, leukemic, and non-leukemic groups and which loadings (PCs) presented statistically significant differences in their intensities between the groups. Chemoface software (version 1.65, UFLA, MG, Brazil, www.ufla.br/chemoface/) [41] was used to perform the PCA.
The PCs were also evaluated for statistical purposes. Gaussian distribution was checked, and ANOVA (Tukey–Kramer) or Kruskal–Wallis (Dunn’s) test was used; differences were considered significant when the p value < 0.05. The p value was used to accept (p value > 0.05) or reject (p value < 0.05) the null hypothesis, i.e., the equality in the intensity of the PCs of normal, leukemic, and non-leukemic groups.
Discriminant analysis by partial least squares (PLS)
Partial least squares discriminant analysis (PLS-DA) is a statistical tool widely used for linear discriminant classification due to its ability to operate with many correlated variables [42,43,44]. The PLS-DA creates a model in which any correlation between the predicted (groups: normal, leukemic, and non-leukemic) and predictor (Raman spectral intensities) variables in the spectral dataset is estimated and maximized by the covariance between the predictor and predicted, and therefore, the model is used to predict the output (classes: normal, leukemic, and non-leukemic) of a new sample, determining which class this sample belongs to. It means that “within groups” variations are distinguished from “between groups” variations, and discrimination is achieved by focusing on “between groups” variations [45].
Chemoface software was used to build the discrimination models with “leave-one-spectrum-out” cross-validation [41]. Two models were used to classify the Raman data: a model in which all the spectra were classified in normal, leukemic, or non-leukemic groups and another model in which spectra were classified in leukemic or non-leukemic groups. With the results obtained from the discrimination models, sensitivity, specificity, and accuracy were calculated for the groups normal, leukemic, and non-leukemic.
Results and discussion
Raman spectroscopy
Human blood serum is an overly complex body fluid comprising a dynamic range of biomolecules such as water, proteins (mainly albumin and alpha- and beta-globulins), amino acids, lipoproteins, hormones, vitamins, glucose, and inorganic salts [33, 46]. It is known that molecular analysis of blood serum can provide information about the metabolic processes that occur in the body, and there is growing interest in using Raman spectroscopy to develop serum-specific sensors for biomolecules for diagnosis and monitoring [32].
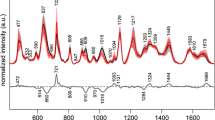
Figure 1 shows the mean Raman spectra of the serum of the normal, leukemic, and non-leukemic groups. The spectra of serum are dominated by features of the main blood serum proteins: albumin and globulins [29, 33], making it difficult to visualize the spectral characteristics of less abundant compounds and small molecules that can function as biomarkers. The peaks of albumin are at the following positions: 828 and 850 cm−1 (tyrosine doublet), 950 cm−1 (C–C stretching mode), and 1002 cm−1 (phenylalanine—δ (= CβH2)4); tryptophan may make a small contribution to the intensity of this band: 1350 cm−1 (CH2 deformation mode), 1450 cm−1 (CH2 deformation mode), and 1650 cm−1 (amide I, alpha-helix conformation). The positions of the Raman peaks of globulins are similar to albumin [29, 36, 47].
Figure 1 also shows the difference spectra between the normal and leukemic groups and between the normal and non-leukemic groups. In the difference spectrum of normal and leukemic groups, the most intense peaks at 622, 1002, 1004, and 1443 cm−1 presented statistically significant differences between groups (ANOVA, p < 0.05). The peak at 645 cm−1 showed statistically significant differences between groups (Kruskal–Wallis, p < 0.05). In the difference spectrum, the peaks at 622 and 1004 cm−1 were more intense in the normal group, and the peaks at 1002 and 1443 cm−1 were more intense in the leukemic group. In the difference spectrum of normal and non-leukemic groups, the most intense peaks at 619, 640, 645, 755, 1165, 1215, and 1446 cm−1 showed statistically significant differences between groups (ANOVA, p < 0.05). The peaks at 622, 1000, 1006, and 1227 cm−1 showed statistically significant differences between groups (Kruskal–Wallis, p < 0.05). The peaks at 622, 645, 1006, 1165, and 1215 cm−1 were more intense in the normal group, and the peaks at 619, 640, 755, 1000, 1227, and 1446 cm−1 were more intense in the non-leukemic group.
Table 2 presents the positions of the main Raman peaks of serum, the biochemical constitution, and the assignments from published literature [20, 27, 33, 34, 36,37,38], as well as the statistical significance (p value) of the peak’s intensities between normal, leukemic, and non-leukemic groups. Significant differences were observed at the following peaks: 643 cm−1 (tyrosine), 717 cm−1 (adenine), 758 cm−1 (tryptophan), 829 cm−1 (tryptophan/tyrosine, glutathione), 1085 cm−1 (tyrosine, phenylalanine), 1178 cm−1 (tyrosine, phenylalanine), 1274 cm−1 (amide III, protein), 1320 cm−1 (tryptophan, phospholipids), 1452 cm−1 (proteins, phospholipids, amino acids), 1557 cm−1 (tryptophan, amide II), and 1659 cm−1 (proteins, alpha-helix, collagen, amide I) (ANOVA test, p < 0.05) and at the peaks: 622 cm−1 (phenylalanine), 1004 cm−1 (phenylalanine, beta-carotene), 1159 cm−1 (beta-carotene), 1209 cm−1 (tryptophan, phenylalanine, adenine, guanine), and 1526 cm−1 (beta-carotene) (Kruskal–Wallis test, p < 0.05); all these peaks in the spectrum of the normal group are more intense when compared to the leukemic and non-leukemic groups. The peaks at 1159 cm−1 (beta-carotene), 1178 cm−1 (tyrosine and phenylalanine), 1209 cm−1 (amide III, tryptophan, phenylalanine, adenine, and thymine), and 1659 cm−1 (phenylalanine) were more intense in the normal group and less intense in the non-leukemic group. The peaks at 1320 cm−1 (tryptophan and phospholipids), 1343 cm−1 (tryptophan, phospholipids, adenine, and guanine), and 1555 cm−1 (tryptophan and amide II) presented higher intensity in the group normal and can differentiate the group normal from the non-leukemic. The peak at 1555 cm−1 was the only one that presented statistical significance between the leukemic and non-leukemic groups (ANOVA test, p < 0.05). The peaks at 852, 901, 943, 1033, 1106, and 1128 cm−1 (ANOVA test, p > 0.05) and the peak 508 cm−1 (Kruskal–Wallis test, p > 0.05) did not show statistically significant differences between the three groups, suggesting that they are not significant for differentiating these groups.
In a study with blood serum samples from healthy and leukemic individuals, González-Solís et al. [20] found that the Raman peaks at 1338 cm−1 (tryptophan, alpha-helix, phospholipids), 1447 cm−1 (lipid), 1523 cm−1 (beta-carotene), 1556 cm−1 (tryptophan), 1587 cm−1 (protein, tyrosine), 1603 cm−1 (tyrosine, phenylalanine), and 1654 cm−1 (proteins— amide I, alpha-helix, phospholipids) were less intense in the spectra of leukemics compared to controls. Minor differences between the leukemic and control groups occurred at 661 cm−1 (glutathione), 890 cm−1 (glutathione), 1126 cm−1 (protein, phospholipid—C–C stretching), 1160 cm−1 (beta-carotene), 1174 cm−1 (tryptophan, phenylalanine), 1208 cm−1 (tryptophan), 1246 cm−1 (amide III), and 1404 cm−1 (glutathione). Increased intensity in the peaks at 973 cm−1 (glucosamine) and 1380 cm−1 (glucosamine) was observed in the leukemic group. The leukemic group also showed a peak at 917 cm−1 (glutathione) that was absent in the control group. The peaks at 1160, 1174, and 1208 cm−1 correspond to the ones in the present study at 1159, 1178, and 1209 cm−1, showing statistically significant increased intensity in the control group.
Silva et al. [27] applied Raman spectroscopy on whole blood and plasma samples to identify spectral differences among healthy subjects and acute leukemic patients based on their biochemical composition (proteins, amino acids, carbohydrates, lipids, and carotenoids). Statistical differences were observed in the peaks at 510 cm−1 (tryptophan), 721 cm−1 (polysaccharides), 760 cm−1 (tyrosine), 837 cm−1 and 947 cm−1 (tyrosine, tryptophan, glutathione), 1004 cm−1 (phenylalanine), 1132 cm−1 (proteins), 1160 cm−1 (beta-carotene), 1210 cm−1 (tryptophan, phenylalanine, tyrosine, amide III), 1269 cm−1 (protein—amide III), 1334 cm−1 (tryptophan), 1344 cm−1 (proteins, tryptophan), 1407 cm−1 (glutathione), 1448 cm−1 (protein), 1455 cm−1 (protein), 1525 cm−1 (beta-carotene), 1630 cm−1 (tyrosine, tryptophan), 1659 cm−1 (phospholipids), and 1666 cm−1 (phospholipids); all peaks in the spectrum of the healthy group were more intense compared to the leukemic group. These peaks were similar to those found in the present study and are attributed to proteins, amino acids, and beta-carotene and are also more intense in the normal (healthy) group compared to the leukemic and non-leukemic groups.
Exploratory analysis
The PCA technique was used to identify the spectral features that presented differences between normal, leukemic, and non-leukemic groups. The PCA extracts the most relevant information (based on the variance) from an original dataset, generating two new principal component variables: SCs and PCs. The SCs represent the axes of spectral variance and resemble Raman spectra, and the PCs indicate the intensities of each loading present in the original data. Such differences may be correlated to the biochemical alterations in blood serum samples from patients with leukemias and other neoplasms (non-leukemic group) compared to normal. The first eight PCA variables are presented in Fig. 2. These variables account for about 98.8% of the total variance observed in the spectral data (96.8%, 0.7%, 0.5%, 0.3%, 0.2%, 0.1%, 0.1%, and 0.05% for SC1 to SC8, respectively).
Plots of the principal component variables PCs and SCs of the Raman serum dataset for exploratory analysis. Superscript letters a, b, and c in the PCs indicate statistical significance (ANOVA test for PC4 and PC6 and Kruskal–Wallis test for PC1, PC2, PC3, PC5, PC7, and PC8 (p < 0.05)) between normal, leukemic, and non-leukemic groups, respectively. NS: not statistically significant
Figure 2 shows the plot of PCs and SCs of serum samples; some PCs showed statistically significant differences between normal, leukemic, and non-leukemic groups. The PC1, PC2, PC3, PC6, and PC7 showed statistically significant differences (ANOVA test, p < 0.05 or Kruskal–Wallis test, p < 0.05), while the PC4, PC5, and PC8 did not show statistically significant differences (ANOVA or Kruskal–Wallis, p > 0.05). According to the literature, the Raman features presented in the first eight SCs were associated with the biochemical compounds of sera. Positive peaks in the SCs with positive PCs suggest that the specific biochemical has a high concentration in that group. Positive peaks with negative PCs suggest that the specific biochemical is with a low concentration in that group.
The SC1 (Fig. 2) showed positive peaks characteristic of serum albumin (852, 1452, and 1659 cm−1) [48, 49]; the PC1 suggested that the serum of normal samples had a higher amount of albumin compared to the serum of leukemic and non-leukemic samples (Dunn’s post-test, normal versus leukemic, p < 0.01; normal versus other types of cancers, p < 0.001). There was no statistically significant difference in the PC1 of the leukemic group versus the non-leukemic group (Dunn’s post-test, normal versus leukemic, p > 0.05), suggesting that such component is not relevant for differentiating the cancer groups. Albumin is the most abundant high molecular weight fraction serum protein in the human body and is synthesized in the liver [49]. The primary physiologic function of albumin is to maintain colloid osmotic pressure, but many other functions have been recognized in the past few years; these include ligand binding and transport of various molecules, in addition to antioxidant and anti-inflammatory actions [29, 50, 51]. Several studies demonstrated that the functions of albumin, such as ligand binding and transport of various molecules, can be applied to the treatment of cirrhotic patients (that can progress to cancer) and patients suffering from other end-stage liver diseases [29]. Individuals with cancer present impairment of defense systems associated with inflammatory processes installed in the body, which culminates in the reduction of plasma albumin [52]. Researchers have shown that low serum albumin levels adversely affect prognosis in cancer [34, 49]. A normal albumin concentration in bodily fluids is considered a sign of good health [29]. The present study corroborates these findings since PC1 suggests that the normal group samples have higher albumin than the leukemic and non-leukemic groups.
In SC2, the “blue shift” (band shift towards high vibrational energy) of the peaks at 1002 cm−1 (inverted peaks at 1002 and 1006 cm−1), 1032 cm−1 (inverted peaks at 1029 and 1034 cm−1), and at 619 and 640 cm−1 (inverted peaks at 619/625 and 640/646 cm−1) was identified in samples from the leukemic and non-leukemic groups compared to normal. In PC2, there is a statistically significant difference between the normal and non-leukemic groups (Dunn’s post-test, p < 0.001) and between the normal and leukemic groups (Dunn’s post-test, p < 0.01), with the data presented by the leukemic group being close to zero. The peaks at 625, 848, and 1002 cm−1 are assigned to phenylalanine [38, 50], an aromatic amino acid with pronounced signature peaks in a Raman spectrum due to the vibration of the benzene ring. The “blue shift” may be due to the conformational alteration of the phenylalanine close to the aromatic ring. The frequencies of the Raman bands belonging to the side chains of the amino acid residues are often blue-shifted concerning the corresponding bands of the free amino acids, especially in the case of the aromatic amino acids [38, 53]. This result suggests that both the variety of protein species and the conformation of proteins changed in serum samples of both leukemic and non-leukemic groups.
SC3 (Fig. 2) showed a negative peak at 1635 cm−1 in the normal group, attributed to amino acids; the positive peaks at 942, 1002, 1308, and 1450 cm−1 are present in the non-leukemic group and are assigned to proteins and amino acids. In PC3, there was a statistically significant difference between the normal and non-leukemic groups (Dunn’s post-test, p < 0.01) and between the leukemic and non-leukemic groups (Dunn’s post-test, p < 0.05); when comparing the normal and leukemic groups, there was no statistically significant difference (Dunn’s post-test, p > 0.05), with the intensity close to zero at the leukemic group. These results may indicate that the conformation and the species variety of proteins are altered in serum samples from cancer patients (non-leukemic group) [53,54,55]. The increase in amino acid levels can be explained by the alteration in the energy metabolism of cancer patients, leading to the synthesis of skeletal muscle and tissue proteins (hepatic gluconeogenesis) and consequent release of amino acids into the blood serum as observed in the samples from the non-leukemic group; this may predispose cancer patients to develop cachexia, compromising their prognosis and quality of life for these patients [55].
The spectral information presented in SC4 (Fig. 2): positive peaks at 624, 645, 1004, 1034, 1134, and 1453 cm−1 and negative peaks at 617, 640, 1002, 1030, and 1650 cm−1 (inverted “blue shift”); SC5: positive peaks at 508, 571, 677, 755, 945, 1004, 1030, 1226, 1341, 1402, 1566, and 1622 cm−1 and negative peaks at 843 and 1440 cm−1, and SC8: positive peaks at 1360, 1504, and 1608 cm−1 and negative peaks at 697, 1004, 1157, 1223, 1435, and 1523 cm−1 are assigned to proteins and amino acids in the samples; although present in different amounts in the serum of the three groups analyzed, these peaks did not show significant differences in the intensities of PC4, PC5, and PC8 (ANOVA test, p = 0.7849; Kruskal–Wallis test, p = 0.0600; Kruskal–Wallis test, p = 0.6082, respectively); therefore, such peaks are not relevant for the differentiation of the groups, but evidencing the heterogeneous and complex nature of the biochemical changes found in the sera of all groups.
The SC6 (Fig. 2) shows positive peaks at 756, 1226, 1304, and 1440 cm−1 and negative peaks at 1003, 1157, 1523, and 1655 cm−1 with a statistically significant difference in the PC6 (ANOVA test, p < 0.05). The positive peaks were assigned to amino acids and proteins and are higher in the non-leukemic group when compared to the normal group (PC6, Tukey–Kramer post-test, p < 0.001); the negative peaks were assigned to carotenoids [20, 33, 56], suggesting that these compounds are higher in the normal group when compared to the non-leukemic group (PC6, Tukey–Kramer post-test, p < 0.001). The group leukemic is close to zero. Higher levels of carotenoids in the blood (serum/plasma) are associated with a decreased risk of cardiovascular disease, heart disease, cancer, and other causes of mortality, as described in other studies [20, 27, 29]. González-Solís et al. [20] used Raman spectroscopy to differentiate normal and leukemic blood serum and to identify the different types of leukemia based on the biochemical compounds of serum. In the normal group (healthy subjects) spectrum, higher amounts of carotenoids were observed (peaks at 1003, 1157, and 1523 cm−1). In a study with blood plasma samples to differentiate between leukemic and healthy subjects, Silva et al. [27] found peak characteristic of carotenoids with significantly higher concentrations in the normal group (1004, 1159/1160, and 1520/1525 cm−1) in the principal component variables SC2/PC2 and SC4/PC4. Such findings agree with those found in the present study.
The SC7 (Fig. 2) showed positive peaks at 502, 619, 640, 1002, 1245, 1412, 1502, and 1683 cm−1 in the normal group and is assigned to proteins and amino acids; the negative peaks at 753, 1006, 1059, 1302, 1453, and 1622 cm−1 are present in the non-leukemic group and are assigned to proteins and amino acids. PC7 showed statistically significant differences between the normal and leukemic groups (Dunn’s post-test, p < 0.001) and between leukemic and non-leukemic groups (Dunn’s post-test, p < 0.001). The positive peaks of proteins (serum albumin—502, 619, 640, and 1002 cm−1) and amino acids present in SC7 are more intense in the normal group, suggesting that these compounds are more frequent in healthy individuals than in cancer individuals (leukemic and non-leukemic groups), corroborating the findings in SC1/PC1.
Discriminant analysis by partial least squares (PLS-DA)
Discriminant analysis using PLS (PLS-DA) was applied to the normalized spectra of the blood serum samples in the normal, leukemic, and non-leukemic groups. PLS-DA models applied to Raman spectral data have been used to classify samples into healthy or neoplastic tissues [27,28,29, 57, 58].
The results of the discrimination models were tabulated in the confusion tables (Table 3), and accuracy, sensitivity, and specificity were calculated. In the PLS-DA discriminating normal, leukemic, and non-leukemic groups (Table 3), maximum accuracy (classification success) of 66% was obtained with six latent variables, with sensitivity of 99% and specificity of 57%. For the discrimination between leukemic and non-leukemic groups (Table 3), the maximum accuracy of 67% was obtained with five latent variables, with sensitivity of 72% and specificity of 60%.
When applying a diagnostic test, there is a possibility of incorrectly classifying individuals, for instance, alleged sick people who are healthy (false positives) and alleged healthy people who are sick (false negatives) [59]. In the proposed study, a sensitivity of 99% was obtained in the differentiation between the normal group (healthy individuals) and leukemic and non-leukemic groups (individuals with cancer) (Table 3). The obtained specificity was 57% (Table 3), which indicates a high rate of false positives. However, diagnostic tests should always be associated with the evaluation of the patient’s clinical history, investigation of the semiology (signs and symptoms present in the physical examination), and results of complementary exams (laboratory, procedural, or imaging) to determine the presence or absence of disease.
The literature available to date is still scarce in comparing patients with different types of neoplasms versus healthy ones through Raman spectroscopy and discriminant analysis in serum, plasma, and blood. Silva et al. [27] used Raman spectroscopy in the blood plasma of healthy and leukemic individuals and obtained, through PLS-DA, accuracy of 97.1%, sensitivity of 95.7%, and specificity of 98%. The cited results are superior to that found in this study. However, the samples used in the present study are more heterogeneous, and the groupings were performed between healthy patients, patients with leukemias (ALL and AML), and patients with other types of cancer, categorized according to the tissue that originated the neoplasia (histology) in sarcoma, blastoma, carcinoma, and lymphoma.
Bai et al. [34] studied the characteristics of blood plasma in patients with diffuse large B-cell lymphoma and chronic lymphocytic leukemia (CLL) by Raman spectroscopy to develop a simple blood test for the non-invasive detection of DLBCL and CLL. The partial orthogonal least squares discriminant analysis (OPLS-DA) was used for discrimination, which can construct two clusters with almost no overlap between DLBCL/CLL and normal. For the LLC model, the sensitivity was 92.9%, and the specificity was 100%, while for the DLBCL model, the sensitivity was 80.0%, and the specificity was 92.3%. Despite using similar statistical analysis to the present study, the models did not group cancer types for discriminant analysis.
Some studies with blood or blood fragments use PCA-DA [20, 32, 33, 38, 60] instead of PLS-DA as a discriminant analysis of the Raman data. However, the PLS-DA stands out for providing information on the differences between groups and recognizing the variances obtained between the groups; therefore, the PLS-DA may lead to better performance in the classification of samples when compared to the PCA-DA [44].
When comparing the Raman spectroscopy with the conventional and currently available techniques for cancer diagnosis, the assessment is performed non-invasively or minimally invasively, without sample preparation, with no need for reagents, and using only a small amount of blood serum [19, 20, 27]. According to studies already available, Raman spectroscopy in blood samples is a promising technique for diagnosis and therapeutic follow-up [19, 20, 27, 33, 57].
The present work reinforces the usefulness of Raman spectroscopy and opens new perspectives for its use to obtain information on biomolecular changes that occur in the human body due to cancer from a blood serum perspective. However, more research is still needed with a larger number of samples, particularly from healthy individuals, to build a spectral database large enough to minimize misclassification, making the data provided by Raman spectroscopy more robust for use in clinical practice. Obtaining more samples will help to investigate the aspects related to the high classification errors that may be due to the heterogeneous nature of biochemical changes in patients with cancer.
Conclusions
In this study, Raman spectroscopy has been applied to blood serum samples from children and adolescents to identify the spectral differences presented in the normal (healthy), leukemic (ALL and AML), and non-leukemic (sarcoma, blastoma, carcinoma, and lymphoma) groups. The mean Raman spectra of the groups showed that the peaks of albumin, proteins, amino acids, and beta-carotene were more intense in the normal group.
Exploratory analysis by PCA applied to the spectra revealed that in SC1, there were characteristic peaks of serum albumin (510, 622, 643, 852, 941, 1003, 1343, 1452, and 1659 cm−1), and PC1 was significantly more intense in the normal group compared to leukemic and non-leukemic groups (Kruskal–Wallis test, p < 0.05); in SC2, a “blue shift” was observed in the peaks at 622 and 643 cm−1 (619/625 cm−1 and 640/646 cm−1), 1003 cm−1 (1002 and 1006 cm−1), and 1032 cm−1 (1029/1034 cm−1) for the serum samples from the leukemic and non-leukemic groups compared to the normal group (Kruskal–Wallis test, p < 0.05); in SC3, positive peaks at 942, 1002, 1308, and 1450 cm−1 (proteins and amino acids) are higher in the non-leukemic group (Kruskal–Wallis test, p < 0.05); in SC6, carotenoids peaks (1003, 1157, and 1523 cm−1) were significantly higher in the normal group compared to the leukemic and non-leukemic groups (ANOVA test, p < 0.05); in SC7, positive peaks at 502, 619, 640, and 1002 cm−1, assigned to serum albumin, were higher in the normal group than in cancer groups (Kruskal–Wallis test, p < 0.05).
The PLS-DA model applied to serum spectra classified normal, leukemic, and non-leukemic groups with accuracy of 66%, sensitivity of 99%, and specificity of 57%. The PLS-DA discriminated leukemic and non-leukemic groups with accuracy of 67%, sensitivity of 72%, and specificity of 60%; the high classification errors may be due to the heterogeneous nature of biochemical changes in patients with cancer and the low number of subjects in the normal group compared to the cancer groups. Raman spectroscopy has shown potential for diagnosing cancer (leukemias and other types of cancer such as sarcomas, lymphomas, blastomas, and carcinomas) through blood serum samples.
References
Allahverdiyev AM, Parlar E, Dinparvar S, Bagirova M, Abamor ES (2018) Current aspects in treatment of breast cancer based of nanodrug delivery systems and future prospects. Artif Cells Nanomed Biotechnol 46:S755–S762. https://doi.org/10.1080/21691401.2018.1511573
Brazil. Ministry of Health. Instituto Nacional de Câncer José Alencar Gomes da Silva (2019) Estimate/2020 - cancer incidence in Brazil. Rio de Janeiro: Instituto Nacional de Câncer José Alencar Gomes da Silva. https://www.inca.gov.br/sites/ufu.sti.inca.local/files//media/document//estimativa-2020-incidencia-de-cancer-no-brasil.pdf. Accessed 5 Jan 2022
Brazil. Ministry of Health (2013) Guidelines for the care of people with chronic diseases in health care networks and in priority care lines - Brasília: Ministry of Health. http://189.28.128.100/dab/docs/geral/documento_norteador.pdf. Accessed 5 Jan 2022
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Global BF (2021) Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21660
Oliveira LS (2021) Childhood cancer: the impact of diagnosis for children and families. REASE 7:2675–3375. https://doi.org/10.51891/rease.v7i5.1223
World Health Organization. Joint WHO/FAO expert consultation on diet, nutrition and the prevention of chronic diseases (2003) Diet, nutrition and the prevention of chronic diseases: report of a joint WHO/FAO expert consultation. WHO Technical Report series 916. Geneva: World Health Organization. http://apps.who.int/iris/bitstream/handle/10665/42665/WHO_TRS_916.pdf;jsessionid=6DCD5B9B3A3347ED819B110F854A184D?sequence=1. Accessed 5 Jan 2022
Brazil. Ministry of Health (2021) Manual of oncology technical bases - SIA/SUS - ambulatory information system. Brasília: Ministry of Health. http://portal.saude.gov.br/portal/saude/Gestor/area.cfm?id_area=1529. Accessed 5 Jan 2022
Manero GG, Chien KS, Bravo GM (2020) Myelodysplastic syndromes: 2021 update on diagnosis, risk stratification and management. Am J Hematol 95:1399–1420. https://doi.org/10.1002/ajh.25950
Brazil. Ministry of Health (2017) Early diagnosis protocol for pediatric oncology. Brasília: Ministry of Health. https://bvsms.saude.gov.br/bvs/publicacoes/protocolo_diagnostico_precoce_cancer_pediatrico.pdf. Accessed 7 Jan 2022
Hanlon EB, Manoharan R, Koo TW, Shafer KE, Motz JT, Fitzmaurice M, Kramer JR, Itzkan I, Dasari RR, Feld MS (2000) Prospects for in vivo Raman spectroscopy. Phys Med Biol 45:R1–R59. https://doi.org/10.1088/0031-9155/45/2/201
Kong K, Kendall C, Stone N, Notingher I (2015) Raman spectroscopy for medical diagnostics - from in-vitro biofluid assays to in-vivo cancer detection. Adv Drug Deliv Rev 89:121–134
Feng S, Huang S, Lin D, Chen G, Xu Y, Li Y, Huang Z, Pan J, Chen R, Zeng H (2015) Surface-enhanced Raman spectroscopy of saliva proteins for the non-invasive differentiation of benign and malignant breast tumors. Int J Nanomedicine 10:537–547. https://doi.org/10.2147/IJN.S71811
Silveira L, Sathaiah S, Zângaro RA, Pacheco MT, Chavantes MC, Pasqualucci CA (2002) Correlation between near-infrared Raman spectroscopy and the histopathological analysis of atherosclerosis in human coronary arteries. Lasers Surg Med 30:290–297. https://doi.org/10.1002/lsm.10053
Bodanese B, Silveira L, Albertini R, Zângaro RA, Pacheco MT (2010) Differentiating normal and basal cell carcinoma human skin tissues in vitro using dispersive Raman spectroscopy: a comparison between principal components analysis and simplified biochemical models. Photomed Laser Surg 28:S119–S127. https://doi.org/10.1089/pho.2009.2565
Lualdi M, Colombo A, Leo E, Morelli D, Vannelli A, Battaglia L, Poiasina E, Marchesini R (2007) Natural fluorescence spectroscopy of human blood plasma in the diagnosis of colorectal cancer: feasibility study and preliminary results. Tumori 6:567–571. https://doi.org/10.1177/030089160709300609
Zhu Y, Fearn T, Chicken DW, Austwick MR, Somasundaram SK, Mosse CA, Clark B, Bigio IJ, Keshtgar MRS, Bown SG (2018) Elastic scattering spectroscopy for early detection of breast cancer: partially supervised Bayesian image classification of scanned sentinel lymph nodes. J Biomed Opt 8:1–9. https://doi.org/10.1117/1.JBO.23.8.085004
Silveira L Jr, Silveira FL, Bodanese B, Zângaro RA, Pacheco MT (2012) Discriminating model for diagnosis of basal cell carcinoma and melanoma in vitro based on the Raman spectra of selected biochemicals. J Biomed Opt 7:077003. https://doi.org/10.1117/1.JBO.17.7.077003
Eberhardt K, Stiebing C, Matthäus C, Schmitt M, Popp J (2015) Advantages and limitations of Raman spectroscopy for molecular diagnostics: an update. Expert Rev Mol 15:773–787. https://doi.org/10.1586/14737159.2015.1036744
Lima AMF, Daniel CR, Navarro RS, Bodanese B, Postulancy CA, Pacheco MTT, Zângaro RA, Silveira L (2019) Discrimination of non-melanoma skin cancer and keratosis from normal skin tissue in vivo and ex vivo by Raman spectroscopy. Vib Spectrosc 15:773–787. https://doi.org/10.1016/j.vibspec.2018.11.009
González-Solís JL, Martínez-Espinosa JC, Salgado-Román JM, Palomares-Anda P (2014) Monitoring of chemotherapy leukemia treatment using Raman spectroscopy and principal component analysis. Lasers Med Sci 29(3):1241–1249. https://doi.org/10.1007/s10103-013-1515-y.E
Kast RE, Tucker SC, Killian K, Trexler M, Honn KV, Auner GW (2014) Emerging technology: applications of Raman spectroscopy for prostate cancer. Cancer Metastasis Rev 33:673–679. https://doi.org/10.1007/s10555-013-9489-6
Plante A, Dallaire F, Groset AA, Nguyen T, Birlea M, Wong J, Daoust F, Roy N, Kougioumoutzakis A, Azzi F, Aubertin K, Kadoury S, Latour M, Albadine R, Prendeville S, Boutros P, Fraser M, Bristow RG, van der Kwast T, Orain M, Brison H, Benzerdjeb N, Hovington H, Bergeron A, Fradet Y, Têtu B, Saad F, Trudel D, Leblond F (2021) Dimensional reduction based on peak fitting of Raman microspectroscopy data improves detection of prostate cancer in tissue specimens. J Biomed Opt 26:116501. https://doi.org/10.1117/1.JBO.26.11.116501
Ikeda H, Ito H, Hikita M, Yamaguchi N, Uragami N, Yokoyama N, Hirota Y, Kushima M, Ajioka Y, Inoue H (2018) Raman spectroscopy for the diagnosis of unlabeled and unstained histopathological tissue specimens. World J Gastrointest Oncol 10:439–448. https://doi.org/10.4251/wjgo.v10.i11.439
Bahreini M, Hoseinzadegan A, Rashidi A, Miri SR, Mirzaei HR, Hajian PA (2019) Raman-based serum constituents analysis for gastric cancer diagnosis: in vitro study. Talanta 204:826–832. https://doi.org/10.1016/j.talanta.2019.06.068
Lazaro-Pacheco D, Shaaban AM, Titiloye NA, Rehman S, Rehman IU (2021) Elucidating the chemical and structural composition of breast cancer using Raman micro-spectroscopy. EXCLI J 20:1118–1132. https://doi.org/10.17179/excli2021-3962
Silveira FL, Pacheco MT, Bodanese B, Pasqualucci CA, Zângaro RA, Silveira L (2015) Discrimination of non-melanoma skin lesions from non-tumor human skin tissues in vivo using Raman spectroscopy and multivariate statistics. Lasers Surg Med 47:6–16. https://doi.org/10.1002/lsm.22318
Silva AM, Siqueira E, Oliveira FSA, Brito PL, Silveira L (2018) Spectral model for diagnosis of acute leukemias in whole blood and plasma through Raman spectroscopy. J Biomed Opt 23:1–11. https://doi.org/10.1117/1.JBO.23.10.107002
Barroso EM, Smits RW, Bakker Schut TC, ten Hove I, Hardillo JA, Wolvius EB, Baatenburg de Jong RJ, Koljenović S, Puppels GJ (2015) Discrimination between oral cancer and healthy tissue based on water content determined by Raman spectroscopy. Anal Chem 87:2419–2426. https://doi.org/10.1021/ac504362y
Parachalil DR, Bruno C, Bonnier F, Blasco H, Chourpa I, McIntyrea J, Byrne HJ (2019) Raman spectroscopic screening of high and low molecular weight fractions of human serum. Analyst 144:4295–4311. https://doi.org/10.1039/c9an00599d
Mitchell K, Ulrich S (2020) Targeting immunophenotypic markers on leukemic stem cells: how lessons from current approaches and advances in the leukemia stem cell (LSC) model can inform better strategies for treating acute myeloid leukemia (AML) cold spring harbor perspectives in medicine. Cold Spring Harb Perspect Med 10:a036251. https://doi.org/10.1101/cshperspect.a036251
Goheen SC, Gilman TH, Kauffman JW, Garvin JE (1977) The effect on Raman spectra of extraction of peripheral proteins from human erythrocyte membranes. Biochem Biophys Res Commun 79:805–814. https://doi.org/10.1016/0006-291X(77)91183-4
Atkins CG, Buckley K, Blades MW, Turner RFB (2017) Raman spectroscopy of blood and blood components. Appl Spectrosc 71(5):767–793. https://doi.org/10.1177/0003702816686593
Martínez-Espinosa JC, González-Solís JL, Frausto-Reyes C, Miranda-Beltrán ML, Soria-Fregoso C, Medina-Valtierra J (2008) Detection of leukemia with blood samples using Raman spectroscopy and multivariate. AIP 1142:99–103. https://doi.org/10.1063/1.3175637
Bai Y, Yu Z, Yi S, Yan Y, Huang Z, Qiu L (2020) Raman spectroscopy-based biomarker screening by studying the fingerprint characteristics of chronic lymphocytic leukemia and diffuse large B-cell lymphoma. J Pharm Biomed Anal 190:133514. https://doi.org/10.1016/j.jpba.2020.113514
Lasch P (2012) Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging. Chemometr Intell Lab 117:100–114. https://doi.org/10.1016/j.chemolab.2012.03.011
Anastasia A, Dmitry A, Bratchenko I (2017) Analysis of albumin Raman scattering registration efficiency from different volume and shape cuvette. J Biomed Photonics Eng 3:020309. https://doi.org/10.18287/JBPE17.03.020309
Hu S, Smith KM, Spiro TG (1996) Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J Am Chem Soc 118:12638–12646. https://doi.org/10.1021/ja962239e
Sjöberg B, Foley S, Cardey B, Enescu M (2014) An experimental and theoretical study of the amino acid side chain Raman bands in proteins. Spectrochim Acta A Mol Biomol Spectrosc 128:300–311. https://doi.org/10.1016/j.saa.2014.02.080
Ringnér M (2008) What is principal component analysis? Nat Biotechnol 26:303–314. https://doi.org/10.1038/nbt0308-303
Jolliffe IT, Cadima J (2016) Principal component analysis: a review and recent developments. Phil Trans R Soc 374:1471–2962. https://doi.org/10.1098/rsta.2015.0202
Nunes AC, Freitas MP, Pinheiro AB, Bastos SC (2012) Chemoface: a novel free user-friendly interface for chemometrics. J Braz Chem 23:2003–2010. https://doi.org/10.1590/S0103-50532012005000073
Ståhle L, Wold S (1987) Partial least squares analysis with cross-validation for the two-clas problem: a Monte Carlo study. J Chemometr 1:185–196. https://doi.org/10.1002/cem.1180010306
Barker M, Rayens W (2003) Partial least squares for discrimination. J Chemometr 17:166–173. https://doi.org/10.1002/cem.785
Bevilacqua M, Marini F (2014) Local classification: locally weighted-partial least squares-discriminant analysis (LW-PLS-DA). Anal Chim Acta 838:20–30. https://doi.org/10.1016/j.aca.2014.05.057
Andries JPM, Vander YH (2021) Improved multi-clas discrimination by common-subset-of-independent-variables partial-least-squares discriminant analysis. Talanta 234:122595. https://doi.org/10.1016/j.talanta.2021.122595
Mescher A (2016) Junqueira’s basic histology text & atlas. McGraw-Hill Medical, New York
Dingari NC, Horowitz GL, Kang JW, Dasari RR, Barman I (2012) Raman spectroscopy provides a powerful diagnostic tool for accurate determination of albumin glycation. PLoS ONE 7:e32406. https://doi.org/10.1371/journal.pone.0032406
Saha A, Yakovlev VV (2010) Structural changes of human serum albumin in response to a low concentration of heavy ions. J Biophotonics 3:670–677. https://doi.org/10.1002/jbio.201000044
Ivanov AI, Korolenko EA, Korolik EV, Firsov SP, Zhbankov RG, Marchewka MK, Ratajczak H (2002) Chronic liver and renal diseases differently affect structure of human serum albumin. Arch Biochem Biophys 408:69–77. https://doi.org/10.1016/S0003-9861(02)00533-7
Arroyo V (2002) Review article: albumin in the treatment of liver diseases–new features of a classical treatment. Aliment Pharmacol Ther 16:1–5. https://doi.org/10.1046/j.1365-2036.16.s5.1.x
Lee JS (2012) Albumin for end-stage liver disease. Korean J Intern Med 27:13–19. https://doi.org/10.3904/kjim.2012.27.1.13
Tang HN, Pan BH, Wang L, Zhu HY, Fan L, Xu W, Li JY (2021) C-reactive protein-to-albumin ratio is an independent poor prognostic factor in newly diagnosed chronic lymphocytic leukaemia: a clinical analysis of 322 cases. Transl Oncol 14:101098. https://doi.org/10.1016/j.tranon.2021.101035
Chen Y, Dai J, Zhou X, Liu Y, Zhang W, Peng G (2014) Raman spectroscopy analysis of the biochemical characteristics of molecules associated with the malignant transformation of gastric mucosa. PLoS One 9:e93906. https://doi.org/10.1371/journal.pone.0093906
Vanna R, Tresoldi C, Ronchi P, Lenferink ATM, Moraso C, Mehn D, Bedoni M, Terstappen LWMM, Ciceri F, Otto C, Gramatica F (2014) Raman spectroscopy for the assessment of acute myeloid leukemia: a proof of concept study. Prog Biomed Opt Imaging 8939:89390F. https://doi.org/10.1117/12.2039186
Sandri M (2016) Protein breakdown in cancer cachexia. Semin Cell Dev Biol 54:11–19. https://doi.org/10.1016/j.semcdb.2015.11.002
Bankapur A, Zachariah E, Chidangil S, Valiathan M, Mathur D (2010) Raman tweezers spectroscopy of live single red and white blood cells. PLoS ONE 5:e10427. https://doi.org/10.1371/journal.pone.0010427
Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller HKH, Vardiman J (2000) Lymphoma classification - from controversy to consensus: the REAL and WHO classification of lymphoid neoplasms. Ann Oncol 11:3–10. https://doi.org/10.1093/annonc/11.suppl_1.S3
Nguyen DV, Rocke DM (2002) Multi-clas cancer classification via partial least squares with gene expression profiles. Bioinformatics 18:1216–1226. https://doi.org/10.1093/bioinformatics/18.9.1216
Pérez I, Vicenti IYT, Xuriguera CGG, Carvajal C, Franco JVA, Loézar C (2021) How to interpret diagnostic tests. Medwave 21:e8432. https://doi.org/10.5867/medwave.2021.07.8432
Managò S, Mirabelli P, Napolitano M, Zito G, de Luca AC (2018) Raman detection and identification of normal and leukemic hematopoietic cells. J Biophotonics 11(5):e201700265. https://doi.org/10.1002/jbio.201700265
Acknowledgements
L. Silveira Jr. acknowledges the partial support of FAPESP (São Paulo Research Foundation) for granting the Raman spectrometer (Process No. 2009/01788-5), CNPq (National Council for Scientific and Technological Development) for the productivity fellowship (Process No. 314167/2021-8), and Ânima Institute (AI) for the research fellowship. A. M. F. Lima and C. R. Daniel acknowledge the support of CAPES (Coordination for the Improvement of Higher Education Personnel) for the doctoral fellowship (Finance Code 001).
Funding
This study has been partly supported by FAPESP (São Paulo Research Foundation, Brazil), which granted the Raman spectrometer (Grant No. 2009/01788–5).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Ana Mara Ferreira de Lima, Camila Ribeiro Daniel, Pedro Luiz de Brito, and Landulfo Silveira Jr performed material preparation, data collection, and analysis. Ana Mara Ferreira de Lima wrote the first draft of the manuscript, and all authors commented on previous versions. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article was approved by the Research Ethics Committee of University Anhembi Morumbi (UAM) (Process CAAE No. 23457019.9.0000.5492) following the Brazilian guidelines for research involving humans. All patients agreed to participate in the research and signed a Free and Informed Consent Term (FICT).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lima, A.M.F., Daniel, C.R., Pacheco, M.T.T. et al. Discrimination of leukemias and non-leukemic cancers in blood serum samples of children and adolescents using a Raman spectral model. Lasers Med Sci 38, 22 (2023). https://doi.org/10.1007/s10103-022-03681-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10103-022-03681-2