Abstract
Objective
We studied genetic risk factors associated with sarcoidosis within a family with a high prevalence of this disease.
Methods
We studied 41 members of a family with a high rate of sarcoidosis, including an index patient with treatment-resistant neurosarcoidosis. Whole genome sequencing was performed for six affected family members and variations associated with loss of function were filtered out as candidate genes. Findings were validated by using amplicon sequencing within all 41 family members with DNA available and candidate genes were screened on absence and presence within the sarcoidosis affected and non-affected.
Results
Family members (n = 61) from 5 generations were available for participation including 13 subjects diagnosed with sarcoidosis (20%). Analyses identified 36 candidate variants within 34 candidate genes. Variations within three of these genes (JAK2, BACH2, and NCF1) previously have been associated with autoimmune diseases.
Conclusions
We identified 34 genes with a possible role in the etiology of sarcoidosis, including JAK2. Our results may suggest evaluation of JAK inhibitors in treatment-resistant sarcoidosis.
Key Points • JAK2 has a potential role in the etiology of sarcoidosis and is a potential therapeutic target. • We identified 33 additional candidate genes of which BACH2 and NCF1 have been previously associated with autoimmune disease. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcoidosis is a granulomatous inflammatory disorder that manifests most commonly between the age of 20 and 40 years [1]. This disease has a prevalence between 5 and 64 per 100,000 with highest incidences in the Northern European and African-American populations [1]. Neurosarcoidosis is a severe variant of sarcoidosis, characterized by granulomas involving the nervous system, with heterogeneous clinical presentation and a high relapse rate [2].
The etiology of sarcoidosis is unknown and it is hypothesized to be caused by an exaggerated immune response to unknown antigens in genetically predisposed individuals [1]. Granuloma formation may be initiated by the innate immune system with a possible role for toll-like receptor 2 [3]. Presentation of antigens through MHC class I or II molecules induces adaptive effector T cell responses, mainly a T helper cell 1 (Th1) and T helper cell 17 response via interleukin 17 (IL-17) and 23 (IL-23) [4]. A genetic basis was shown by a twin study, estimating a heritability of 66% [5]. Genome-wide association studies (GWAS) and candidate gene studies have described candidate genes, including annexin A11 (ANXA11), butyrophilin-like 2 (BTNL2), coiled-coil domain containing 88B (CCDC88B), BCL2-associated agonist of cell death (BAD), and associations in the human leukocyte antigen (HLA), most notably HLA-DRB1 and HLA-DBQ1 [4].
At our outpatient clinic, a tertiary referral center for sarcoidosis, we lost a patient with treatment refractory neurosarcoidosis due to a relapse of the disease. Our patient belonged to a Suriname family with an unusual high rate of sarcoidosis. At the request of our patient and her family, we tried to unravel genetic risk factors associated with sarcoidosis using whole genome sequencing.
Methods
We constructed a pedigree based on the clinical information provided by the family. Not all information of the fourth and fifth generations could be provided. Diagnosis of sarcoidosis was made by the treating physician, according to international guidelines [1]. Tissue confirmation of non-caseating granulomatous inflammation was preferred but not required for inclusion. We collected data on medication use, diagnosis, organ involvement, treatment response, and outcome. This study was done at the request of the family. All participants provided written informed consent and the study was done according to Dutch legislation.
Saliva of 40 family members was collected for DNA extraction and isolated using the Oragene saliva collection system (OG-500, DNA Genotek) according to manufacturer’s protocol. Additionally, blood was withdrawn from one patient in sodium/EDTA tubes for DNA extraction. Isolation of DNA was performed with the Gentra Puregene isolation kit (QIAGEN) according to the manufacturer’s protocol.
For the whole genome sequencing, DNA concentration was determined by means of fluorometric measurement (Qubit, Thermi) and quality was checked by determining the absence of degradation and presence of high molecular weight DNA. Whole genome sequencing of six sarcoidosis-affected family members was done using the Illumina HiSeq X platform at the Hartwig Medical Foundation DNA sequencing center. Paired-end reads for each individual were merged using Picard (version 1.92) and aligned to the GRch37/HG19 reference genome using the LifeScope aligner (version 2.5.1, Applied Biosystems). To minimize mismatched bases between reads, realignments were performed using the RealignerTargetCreator function in GATK (version 2.7-4). Samples were recalibrated with GATK Recalibrate and variants were called using GATKs HaplotypeCaller (version 3.3-0) with default settings. Variants for analysis were filtered on a minimal read depth of 20 and genotype quality of 99. After filtering, all samples were combined and genotyped by using GATK CombineGVCFs and GenotypeGVCFs (version 3.3-0). Finally, all chromosomal locations for found variants were annotated using SnpEff and the UCSC variant annotation integrator tool (https://genome.ucsc.edu/) [6]. We updated all mentioned locations in the manuscript according to the GRCh38 using Lift Genome Annotations for the UCSC genome browser.
For the validation of candidate variants, AmpliSeqؑ™ (Life Technologies, Carlsbad, CA, USA) custom panels were developed using Ion AmpliSeq™ designer software capturing 307 candidate variants. Libraries were constructed using Ion AmpliSeq™ Library Kit (v2.0, Thermo Fisher) according to the manufacturer’s instructions. Validation and quantification of enriched targeted DNA were performed. Emulsion PCR was performed using the SOLiD EZ Bead Emulsifier and Amplifier (Applied Biosystems). Sequencing was performed on the SOLiD 5500xl sequencer (Life Technologies) generating paired-end reads (50 bp forward and 35 bp reverse).
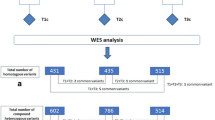
In the analysis pipeline, we used the six whole genome sequenced affected family members as a template to find variations associated with loss of function of a gene, consisting of variants with frameshift, in-frame indel, start/stop codon change, and missense using Ingenuity Variant Analysis™ (QIAGEN Redwood City, CA). All exomic variants that had a call quality of more than 20 were imported in Ingenuity Variant Analysis™. Filters were set to exclude common variants, remove variants in the top 5% most exonically variable and all variants with an allele frequency of ≥ 1% in the 1000 Genomes, ExAC, and NHLBI ESP databases. We kept variants which were associated with gain of function or which were homozygous, haplo-insufficient, or hemizygous.
Using AmpliSeq™, we verified the Ingenuity Variant Analysis™ results for all six affected family members and 35 other family members for whom we were able to collect DNA. Overall, we performed amplicon sequencing in 41 individuals, including the six whole genome sequenced individuals. Two researchers independently evaluated all AmpliSeq™ results by visually inspecting the reads using Integrative Genomics Viewer (IGV, Broad Institute, Cambridge, MA, USA). Low-quality variants and identical variation between all family members were excluded. Protein function and/or allele frequency difference between affected and non-affected family members was used to further select variations. An independent in-house reference whole exome sequencing database was used to exclude all variants present with a minor allele frequency (MAF) of 0.01. Finally, we manually re-evaluated all population MAF of the remaining variations by consulting ExAC. Furthermore, all variants were evaluated using the combined annotation-dependent depletion (CADD) score and the variants were cross referenced in the ClinVar database.
All remaining genes were thereafter evaluated on function and previously reported roles in sarcoidosis, other granulomatous and/or autoimmune disease, or a role in the immune system. Furthermore, we performed pathway analysis using the online tool from the STRING database. STRING database was used to evaluate if the reported proteins in which our variants were found had any similar interaction networks or functional enrichment analysis.
Data availability statement
Anonymized data not published in the article is available on request by any qualified investigator.
Results
Family members from 5 generations were available for participation in the study (Fig. 1), consisting of 61 subjects of whom 13 were diagnosed with sarcoidosis (21%). Of these, genetic material could be collected from 41 patients (67%). The family was of African Surinamese ancestry. Characteristics of the 13 affected patients are presented in Table 1. The diagnosis of sarcoidosis was confirmed by pathological examination in 8 of the 13 patients (62%), based on clinical suspicion in combination with imaging in 4 patients and on clinical suspicion only in 1 patient. The latter was a patient with an isolated eye manifestation. Organ system involvement consisted of the lungs in 11 patients, eyes in 3, joints in 3, skin in 2, and nervous system in 2. Treatment was known for 8 patients and consisted of systemic use of immunosuppressive therapy in 5 patients as follows: prednisone in 5, methotrexate in 2, and infliximab, chloroquine, and azathioprine in 1 patient. One patient was treated with prednisolone eye drops only. Treatment response was known for 8 of 13 patients and consisted of complete remission in 7 (88%) and death due to sepsis in the index patient.
Whole genome sequencing was done on the 6 patients with sarcoidosis (Fig. 1). These patients had various affected organ systems, including the nervous system (in 2 patients, one of which is the index patient), lungs (5 patients), eyes (3 patients), liver (1 patient), and skin (2 patients). All but one had pathology-confirmed sarcoidosis. Ingenuity Variant Analysis™ filtering resulted in 358 candidate variants within 132 different genes in the six affected family members with a potential association with development of sarcoidosis. Supplementary figure 1 shows our step-by-step workflow of selection of candidate genes. We developed primers for AmpliSeq™ to sequence and evaluated 294 of the candidate variations leading to 36 variants within 34 genes passing the initial filtering (Table 2, Supplementary figure 1). Pathway analysis showed no associations of the 34 genes and overrepresentation of a specific pathway. This means that no general pathway within the reported relevant variations is detected to explain the disease cause. The genes with the highest minor allele frequency (MAF) in affected versus unaffected patients were ZFAT (MAF affected/unaffected, 4.57), DNAH9 (MAF affected/unaffected, 3.43), NCF1 (MAF affected/unaffected, 3.43), and MCM2 (MAF affected/unaffected, 3.24). The genes with the highest CADD scores were OTOG (28.5), MYF6 (27.7), and NUN6 (26.6). In total, 5 genes had a CADD score below 1, consisting of SETSIP, EHMT1, MUC5B, CSH1/CSH2, and BIRC5. Furthermore, evaluation with the ClinVar database did not result in additional information.
From the 34 discovered genes, 14 have previously been described to have a role in the immune system. A total of 6 genes are involved in the adaptive immune system, including T and B cell activation or proliferation (RGS2, AGRN, ETS1, IGF2R, SMO, and ZFAT) and 3 have a role in the innate immune system, including dendrocyte and/or macrophage activation or migration (POLR3A, NR1H3, and FLT1) [7, 8]. Additionally, BACH2 codes for a transcription factor involved in both the macrophage-mediated innate immune response and the adaptive immune response [9]. Furthermore, four genes have various roles directly linked to the immune system. CR1L is involved in the complement system (CR1L) [10]. NCF1 and the x-chromosomal CYBB code for a subunit of neutrophil nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, which is a membrane-bound enzyme complex that generates superoxide [11]. JAK2 codes for a protein tyrosine kinase, which is required for responses to interferon gamma (IFNγ), granulocyte-macrophage colony-stimulating factor (GM-CSF), and several interleukins (IL-3, IL-5, IL-6, and IL-12, among others) [12].
Of the remaining 20 genes, 18 genes have functions that have not been associated with the immune system and two genes have an unknown function. Six genes are involved in the cardiovascular system (SETSIP), respiratory tract (DNAH9), digestive tract (MALRD1), ear and nose (OTOG and OR11H12), and musculoskeletal system (MYF6) [13,14,15,16,17]. Additionally, DSP codes for desmoplakin and is involved in cell structure in cardiac tissue and epidermal cells and MUC5B codes for the components of mucus and is mainly expressed in the lungs [18, 19]. Ten genes have various roles in the cell cycle, mainly cell division and proliferation (FRY, BIRC5, CHD7, and EHMT1), but also DNA replication, cell cycle regulatory proteins and genomic stability, and repair (CDK12, SPRY4, and MCM2, respectively) [20, 21]. Also, two genes have roles in RNA modification and translation initiation (NSUN6 and PABPC3) and CSH1 codes for a protein that is only secreted during pregnancy and stimulates fetal growth [22, 23]. Of the remaining two genes, the exact function is unknown (MYO15B and FAM166A).
Discussion
We identified 36 variants in 34 genes with possible roles associated with sarcoidosis in a large family with a high prevalence of the disease. We identified genetic variations in JAK2, BACH2, and NCF1 that have been associated with autoimmune diseases such as inflammatory bowel disease and chronic granulomatous disease [24, 25].
Identification of the JAK/STAT pathway is of special interest because of potential clinical use of JAK-specific inhibitors in sarcoidosis [26]. JAK2 codes for a protein tyrosine kinase, which is required for responses to interferon gamma (IFNγ), granulocyte-macrophage colony-stimulating factor (GM-CSF), and several interleukins (IL-3, IL-5, IL-6, and IL-12, among others) [12]. Both IFNγ and GM-CSF have been associated with sarcoidosis and disease activity [27, 28]. Upon activation, JAK2 phosphorylates signal transducer and activator of transcription (STAT) and initiates the JAK/STAT signaling pathway. The JAK/STAT pathway has been implicated in both the IL-12 and IL-23 signaling pathways, which leads to Th1 and Th17 CD4 cell maturation [12]. These Th cells are important mediators of immune responses and are thought to organize the granulomatous structure, which is in turn highlighted by CD4 T cell lymphopenia in sarcoidosis patients [29]. In a study evaluating microRNA expression and protein-coding gene expression in sarcoidosis patients, they found the JAK/STAT signaling pathway to be the most significantly involved pathway [30]. A genetic association study involving 1996 German sarcoidosis patients described an overlap between risk loci in inflammatory bowel disease and sarcoidosis, especially in the IL-23 signaling pathway [31]. Authors described two variants in the JAK2 gene associated with chronic sarcoidosis [31].
JAK is a novel therapeutic target in inflammatory bowel disease. The two major forms of inflammatory bowel disease, Crohn’s disease and ulcerative colitis, are also chronic immune-mediated conditions characterized by an increased production of pro-inflammatory cytokines leading to granuloma formation [26]. In line with our findings in sarcoidosis patients, genetic association studies have linked inflammatory bowel disease to the JAK/STAT pathway [32]. In two randomized clinical trials, involving patients with moderately to severely active ulcerative colitis, tofacitinib was more effective as induction and maintenance therapy than a placebo [26]. Tofacitinib, a pan-JAK inhibitor, has recently been approved for the treatment of moderate-to-severe ulcerative colitis [33]. Furthermore, positive effects of tofacitinib have been described in patients with sarcoidosis [34]. We previously described that one-third of patients with neurosarcoidosis do not respond to treatment [2]. Future research might evaluate JAK inhibitors in patients with treatment-resistant sarcoidosis.
BACH2 codes for a transcription factor involved in both the macrophage-mediated innate immune response and the adaptive immune response. It is expressed in B and T cells, alveolar macrophages, and neural cells [9]. BACH2 is a broad regulator of immune activation and is required for the formation of regulatory T cells. It has an essential role in maintaining immune homeostasis [9,3 5]. Furthermore, it limits the Th1 and Th17 differentiation to effector lineages CD4+ T cells [35]. In mice knocked out for BACH2, investigators found an increase in the number of CD4 T cells in the lungs and peripheral lymphoid organs and increased proportions of effector cells in the lungs. As described above, these CD4 T cells play an important role in granuloma organization. Most notably, variations in the BACH2 gene are associated with Crohn’s disease [24]. Only one study found a possible role for BACH2 in sarcoidosis patients. They found a negative correlation between gene expression of BACH2 and disease severity [30].
Variants in NCF1 were one of the most distinctive variations, which were more frequently found in affected family members. NCF1 codes for a 47-kDa cytosolic subunit of neutrophil NADPH oxidase. Mutations in this gene lead to a lower level of reactive oxygen species leading to decreased intracellular killing in phagocytic cells [11]. NCF1 is associated with chronic granulomatous disease. Patients with this disease have increased susceptibility to recurrent infections and granuloma formation. We have identified an additional candidate gene, the x-chromosomal CYBB, with a common pathway, also associated with this disease. NCF1 mutations have also been associated with an increased susceptibility for systemic lupus erythematosus (SLE), Sjögren’s syndrome, and rheumatoid arthritis [25]. Sarcoidosis in patients with chronic granulomatous disease has been described, but is rare which might be due to underreporting given the difficulty to discriminate between the two diseases [36]. Animal studies have shown that NCF1-mutated mice show an increase of T cell–dependent autoimmunity [37]. Furthermore, NAHDP deficiency may play a role in decreased antigen degradation and increased cross-presentation of antigens via MHC class I or II between dendritic cells, possibly enhancing auto-immunity [38]. In mice with a loss of NAHDP function, an increase of non-caseating granuloma formation was observed in response to Propionibacterium acnes [39]. Previous studies have demonstrated the occurrence of microbes in patients with sarcoidosis leading to the hypothesis that decreased bacterial clearance may add to granuloma formation [40].
Sarcoidosis is thought to be a complex disease in which many genetic variations with small effect are involved in addition to unknown environmental factors [1]. Multiple variants have been found in genome-wide association studies and replication of these results proved to be difficult, as illustrated by HLA-DR9 which has been associated with disease risk in Japanese patients, but with disease protection in Scandinavians [41]. It has been suggested that population stratification and failure to correct for population-specific or ethnicity-specific traits remain a limitation and this might be improved when cohorts are stratified on the basis of genetically determined ancestry [42]. Family-based studies have the capacity to limit the confounding ancestry-specific genetic predisposition. For example, early family linkage studies have linked a familial risk to the MHC region, which have further been pinpointed to haplotype variation in the region of the BTNL2 gene with the strength of the association depending on ethnicity [43]. The advantage of our approach, which is performed within a family with affected individuals with a shared ancestry, is that it can focus on rare genetic triggers with strong effects, keeping in mind the polygenetic nature of the disease. Recently, three family studies were published evaluating genetic predisposition using a comparable approach [44,45,46]. A whole exome sequencing study in 14 affected individuals and 8 unaffected family members in 5 different families used prediction software and a minor allele frequency lower than 0.05 to further select candidate genes and found 227 variants in 192 genes. Analysis of the involved pathways identified the TOR pathway as a possible association. No information on the ethnic background or admixture of the selected families was published. Another whole exome sequencing study in 22 affected German patients from 6 families and 14 from 5 families did not use healthy family members as a control group. They looked at shared genetic predisposition between the families and variants were filtered if they occurred in less than 50% of each pedigree. They found 40 candidate variants involved in Wnt signaling, chemokine- and cytokine-mediated inflammatory responses and cadherin signaling pathways. Interestingly, no commonly shared genomic region or gene was identified among the analyzed six sarcoidosis pedigrees, highlighting the complex and heterogeneous genotype-phenotype relation of sarcoidosis. A third study investigated traits associated with disease onset before the age of 15 in patients and used their healthy parents as controls. They selected variants that only occurred in affected patients or were transmitted as recessive traits from each parent. Ethnic background was not provided. They found 37 candidate genes, but none was shared by all patients. Altogether, many genetic variations are suggested to be of influence in sarcoidosis, but few are replicated in a population with shared ethnic ancestry. Further exploration in multiple families with a shared ancestry with high prevalence of sarcoidosis is an important next step in future studies to identify candidate variants and genes.
Our study has several limitations. First, DNA was collected from a large number of family members but not all. Second, clinical data could not be collected in all patients especially in the first two generations as the diagnosis was made more than 20 years ago, subjects died, or were living abroad. In the individuals of whom data could not be collected, clinical information was verified by family members, which may be unreliable. However, the diagnosis was confirmed by biopsy in 67% of the individuals that limits the diagnostic uncertainty. Third, patients of the fourth and fifth generations may still develop sarcoidosis due to their relatively young age. Fourth, we cannot exclude that there is a non-genetic shared exposure to unknown antigens in addition to a shared genetic risk leading to a bias in our results. However, this cannot be overcome in a family study. The size of the family including different generations living in different parts of The Netherlands and Suriname is expected to limit shared exposure to unknown antigens. Fifth, the presence of variations within other populations is an important filter in our analysis to selected candidate variations associated with sarcoidosis. The ideal control population for this family of Suriname origin would have been one with African-Caribbean descent. With the exception of the Barbados 1000 genome project population, African-Caribbean’s populations are underrepresented in population genetics, which could have affected the selection of associated variations [47]. Currently, no other African-Caribbean sarcoidosis cohorts or families have been described making validation of our results difficult and we could not investigate the role of these variants in the pathogenesis of sarcoidosis. In line with autoimmune disease GWAS, we do not find a single causal gene but multiple candidate genes within the family for sarcoidosis indicating a multifactorial genetic origin for this disease. This indicates the complexity of detecting the involved genetic variability in sarcoidosis risk and possible correlation with the highly variable presentation and progression of the disease [4]. Lastly, IVA does not target certain structural variants and deletions and duplications might be missed in our analysis.
In this family-based study, we have identified 36 variants in 34 candidate genes with a possible role in the etiology of sarcoidosis. Identified variations within three of these genes, namely JAK2, BACH2, and NCF1, have previously been associated with multiple autoimmune diseases. Of special interest is the role of JAK/STAT pathway in the pathophysiology of sarcoidosis. JAK inhibitors have been approved for colitis ulcerosa, a disease with many similar aspects in the pathophysiology, so this may be a novel treatment option for treatment-resistant sarcoidosis.
Data availability
Anonymized data not published in the article is available on request by any qualified investigator.
References
Valeyre D, Prasse A, Nunes H, Uzunhan Y, Brillet PY, Muller-Quernheim J (2014) Sarcoidosis. Lancet 383(9923):1155–1167. https://doi.org/10.1016/S0140-6736(13)60680-7
Fritz D, van de Beek D, Brouwer MC (2016) Clinical features, treatment and outcome in neurosarcoidosis: Systematic review and meta-analysis. BMC Neurol 16(1):220. https://doi.org/10.1186/s12883-016-0741-x
Gabrilovich MI, Walrath J, van Lunteren J, Nethery D, Seifu M, Kern JA, Harding CV, Tuscano L, Lee H, Williams SD, Mackay W, Tomashefski JF Jr, Silver RF (2013) Disordered Toll-like receptor 2 responses in the pathogenesis of pulmonary sarcoidosis. Clin Exp Immunol 173(3):512–522. https://doi.org/10.1111/cei.12138
Fingerlin TE, Hamzeh N, Maier LA (2015) Genetics of sarcoidosis. Clin Chest Med 36(4):569–584. https://doi.org/10.1016/j.ccm.2015.08.002
Sverrild A, Backer V, Kyvik KO, Kaprio J, Milman N, Svendsen CB, Thomsen SF (2008) Heredity in sarcoidosis: A registry-based twin study. Thorax 63(10):894–896. https://doi.org/10.1136/thx.2007.094060
Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. https://doi.org/10.4161/fly.19695
Fisher RA (2015) Preface. RGS protein physiology and pathophysiology. Prog Mol Biol Transl Sci 133:xi–xii. https://doi.org/10.1016/S1877-1173(15)00122-2
Sawano A, Iwai S, Sakurai Y, Ito M, Shitara K, Nakahata T, Shibuya M (2001) Flt-1, vascular endothelial growth factor receptor 1, is a novel cell surface marker for the lineage of monocyte-macrophages in humans. Blood 97(3):785–791
Zhou Y, Wu H, Zhao M, Chang C, Lu Q (2016) The Bach family of transcription factors: A comprehensive review. Clin Rev Allergy Immunol 50(3):345–356. https://doi.org/10.1007/s12016-016-8538-7
Molina H, Wong W, Kinoshita T, Brenner C, Foley S, Holers VM (1992) Distinct receptor and regulatory properties of recombinant mouse complement receptor 1 (CR1) and Crry, the two genetic homologues of human CR1. J Exp Med 175(1):121–129
Kuhns DB, Hsu AP, Sun D, Lau K, Fink D, Griffith P, Huang DW, Priel DAL, Mendez L, Kreuzburg S, Zerbe CS, De Ravin SS, Malech HL, Holland SM, Wu X, Gallin JI (2019) NCF1 (p47(phox))-deficient chronic granulomatous disease: Comprehensive genetic and flow cytometric analysis. Blood Adv 3(2):136–147. https://doi.org/10.1182/bloodadvances.2018023184
Cho JH, Gregersen PK (2011) Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med 365(17):1612–1623. https://doi.org/10.1056/NEJMra1100030
Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai TN, Baban D, Ragoussis J, Huang Y, Han JD, Zeng L, Hu Y, Xu Q (2012) Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci U S A 109(34):13793–13798. https://doi.org/10.1073/pnas.1205526109
Reue K, Lee JM, Vergnes L (2014) Regulation of bile acid homeostasis by the intestinal Diet1-FGF15/19 axis. Curr Opin Lipidol 25(2):140–147. https://doi.org/10.1097/MOL.0000000000000060
Tilley AE, Walters MS, Shaykhiev R, Crystal RG (2015) Cilia dysfunction in lung disease. Annu Rev Physiol 77:379–406. https://doi.org/10.1146/annurev-physiol-021014-071931
Simmler MC, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou JC, Petit C, Panthier JJ (2000) Targeted disruption of otog results in deafness and severe imbalance. Nat Genet 24(2):139–143. https://doi.org/10.1038/72793
Kerst B, Mennerich D, Schuelke M, Stoltenburg-Didinger G, von Moers A, Gossrau R, van Landeghem FK, Speer A, Braun T, Hubner C (2000) Heterozygous myogenic factor 6 mutation associated with myopathy and severe course of Becker muscular dystrophy. Neuromuscul Disord 10(8):572–577
Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, Tuvim MJ, Adachi R, Romo I, Bordt AS, Bowden MG, Sisson JH, Woodruff PG, Thornton DJ, Rousseau K, De la Garza MM, Moghaddam SJ, Karmouty-Quintana H, Blackburn MR, Drouin SM, Davis CW, Terrell KA, Grubb BR, O’Neal WK, Flores SC, Cota-Gomez A, Lozupone CA, Donnelly JM, Watson AM, Hennessy CE, Keith RC, Yang IV, Barthel L, Henson PM, Janssen WJ, Schwartz DA, Boucher RC, Dickey BF, Evans CM (2014) Muc5b is required for airway defence. Nature 505(7483):412–416. https://doi.org/10.1038/nature12807
Favre B, Begre N, Borradori L (2018) A recessive mutation in the DSP gene linked to cardiomyopathy, skin fragility and hair defects impairs the binding of desmoplakin to epidermal keratins and the muscle-specific intermediate filament desmin. Br J Dermatol 179(3):797–799. https://doi.org/10.1111/bjd.16832
Kuo HH, Ahmad R, Lee GQ, Gao C, Chen HR, Ouyang Z, Szucs MJ, Kim D, Tsibris A, Chun TW, Battivelli E, Verdin E, Rosenberg ES, Carr SA, Yu XG, Lichterfeld M (2018) Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4(+) T cells. Immunity 48(6):1183–1194 e1185. https://doi.org/10.1016/j.immuni.2018.04.004
Mason JM, Morrison DJ, Basson MA, Licht JD (2006) Sprouty proteins: Multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 16(1):45–54. https://doi.org/10.1016/j.tcb.2005.11.004
Haag S, Warda AS, Kretschmer J, Gunnigmann MA, Hobartner C, Bohnsack MT (2015) NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21(9):1532–1543. https://doi.org/10.1261/rna.051524.115
Mannik J, Vaas P, Rull K, Teesalu P, Rebane T, Laan M (2010) Differential expression profile of growth hormone/chorionic somatomammotropin genes in placenta of small- and large-for-gestational-age newborns. J Clin Endocrinol Metab 95(5):2433–2442. https://doi.org/10.1210/jc.2010-0023
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 42(12):1118–1125. https://doi.org/10.1038/ng.717
Zhao J, Ma J, Deng Y, Kelly JA, Kim K, Bang SY, Lee HS, Li QZ, Wakeland EK, Qiu R, Liu M, Guo J, Li Z, Tan W, Rasmussen A, Lessard CJ, Sivils KL, Hahn BH, Grossman JM, Kamen DL, Gilkeson GS, Bae SC, Gaffney PM, Shen N, Tsao BP (2017) A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat Genet 49(3):433–437. https://doi.org/10.1038/ng.3782
Danese S, Argollo M, Le Berre C, Peyrin-Biroulet L (2019) JAK selectivity for inflammatory bowel disease treatment: Does it clinically matter? Gut. https://doi.org/10.1136/gutjnl-2019-318448
Itoh A, Yamaguchi E, Furuya K, Hizawa N, Ohnuma N, Kawakami Y, Kuzumaki N (1993) Correlation of GM-CSF mRNA in bronchoalveolar fluid with indices of clinical activity in sarcoidosis. Thorax 48(12):1230–1234
Prior C, Haslam PL (1991) Increased levels of serum interferon-gamma in pulmonary sarcoidosis and relationship with response to corticosteroid therapy. Am Rev Respir Dis 143(1):53–60. https://doi.org/10.1164/ajrccm/143.1.53
Timmermans WM, van Laar JA, van Hagen PM, van Zelm MC (2016) Immunopathogenesis of granulomas in chronic autoinflammatory diseases. Clin Transl Immunol 5(12):e118. https://doi.org/10.1038/cti.2016.75
Zhou T, Casanova N, Pouladi N, Wang T, Lussier Y, Knox KS, Garcia JGN (2017) Identification of Jak-STAT signaling involvement in sarcoidosis severity via a novel microRNA-regulated peripheral blood mononuclear cell gene signature. Sci Rep 7(1):4237. https://doi.org/10.1038/s41598-017-04109-6
Fischer A, Nothnagel M, Franke A, Jacobs G, Saadati HR, Gaede KI, Rosenstiel P, Schurmann M, Muller-Quernheim J, Schreiber S, Hofmann S (2011) Association of inflammatory bowel disease risk loci with sarcoidosis, and its acute and chronic subphenotypes. Eur Respir J 37(3):610–616. https://doi.org/10.1183/09031936.00049410
Lee JC, Biasci D, Roberts R, Gearry RB, Mansfield JC, Ahmad T, Prescott NJ, Satsangi J, Wilson DC, Jostins L, Anderson CA, Consortium UIG, Traherne JA, Lyons PA, Parkes M, Smith KG (2017) Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 49(2):262–268. https://doi.org/10.1038/ng.3755
Tran V, Shammas RM, Sauk JS, Padua D (2019) Evaluating tofacitinib citrate in the treatment of moderate-to-severe active ulcerative colitis: Design development and positioning of therapy. Clin Exp Gastroenterol 12:179–191. https://doi.org/10.2147/CEG.S150908
Damsky W, Thakral D, Emeagwali N, Galan A, King B (2018) Tofacitinib treatment and molecular analysis of cutaneous sarcoidosis. N Engl J Med 379(26):2540–2546. https://doi.org/10.1056/NEJMoa1805958
Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, Sciume G, Zare H, Vahedi G, Dema B, Yu Z, Liu H, Takahashi H, Rao M, Muranski P, Crompton JG, Punkosdy G, Bedognetti D, Wang E, Hoffmann V, Rivera J, Marincola FM, Nakamura A, Sartorelli V, Kanno Y, Gattinoni L, Muto A, Igarashi K, O’Shea JJ, Restifo NP (2013) BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498(7455):506–510. https://doi.org/10.1038/nature12199
De Ravin SS, Naumann N, Robinson MR, Barron KS, Kleiner DE, Ulrick J, Friend J, Anderson VL, Darnell D, Kang EM, Malech HL (2006) Sarcoidosis in chronic granulomatous disease. Pediatrics 117(3):e590–e595. https://doi.org/10.1542/peds.2005-1349
Hultqvist M, Olofsson P, Holmberg J, Backstrom BT, Tordsson J, Holmdahl R (2004) Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc Natl Acad Sci U S A 101(34):12646–12651. https://doi.org/10.1073/pnas.0403831101
Gardiner GJ, Deffit SN, McLetchie S, Perez L, Walline CC, Blum JS (2013) A role for NADPH oxidase in antigen presentation. Front Immunol 4:295. https://doi.org/10.3389/fimmu.2013.00295
Werner JL, Escolero SG, Hewlett JT, Mak TN, Williams BP, Eishi Y, Nunez G (2017) Induction of pulmonary granuloma formation by Propionibacterium acnes is regulated by MyD88 and Nox2. Am J Respir Cell Mol Biol 56(1):121–130. https://doi.org/10.1165/rcmb.2016-0035OC
Chen ES, Moller DR (2015) Etiologies of sarcoidosis. Clin Rev Allergy Immunol 49(1):6–18. https://doi.org/10.1007/s12016-015-8481-z
Berlin M, Fogdell-Hahn A, Olerup O, Eklund A, Grunewald J (1997) HLA-DR predicts the prognosis in Scandinavian patients with pulmonary sarcoidosis. Am J Respir Crit Care Med 156(5):1601–1605. https://doi.org/10.1164/ajrccm.156.5.9704069
Thompson CL, Rybicki BA, Iannuzzi MC, Elston RC, Iyengar SK, Gray-McGuire C, Sarcoidosis Genetic Analysis C (2006) Reduction of sample heterogeneity through use of population substructure: An example from a population of African American families with sarcoidosis. Am J Hum Genet 79(4):606–613. https://doi.org/10.1086/507847
Schurmann M, Lympany PA, Reichel P, Muller-Myhsok B, Wurm K, Schlaak M, Muller-Quernheim J, du Bois RM, Schwinger E (2000) Familial sarcoidosis is linked to the major histocompatibility complex region. Am J Respir Crit Care Med 162(3 Pt 1):861–864. https://doi.org/10.1164/ajrccm.162.3.9901099
Calender A, Rollat Farnier PA, Buisson A, Pinson S, Bentaher A, Lebecque S, Corvol H, Abou Taam R, Houdouin V, Bardel C, Roy P, Devouassoux G, Cottin V, Seve P, Bernaudin JF, Lim CX, Weichhart T, Valeyre D, Pacheco Y, Clement A, Nathan N, in the frame of GSF (2018) Whole exome sequencing in three families segregating a pediatric case of sarcoidosis. BMC Med Genet 11(1):23. https://doi.org/10.1186/s12920-018-0338-x
Calender A, Lim CX, Weichhart T, Buisson A, Besnard V, Rollat-Farnier PA, Bardel C, Roy P, Cottin V, Devouassoux G, Finat A, Pinson S, Lebecque S, Nunes H, Israel-Biet D, Bentaher A, Valeyre D, Pacheco Y, in the frame of GSF (2019) Exome sequencing and pathogenicity-network analysis of five French families implicate mTOR signalling and autophagy in familial sarcoidosis. Eur Respir J 54(2). https://doi.org/10.1183/13993003.00430-2019
Kishore A, Petersen BS, Nutsua M, Muller-Quernheim J, Franke A, Fischer A, Schreiber S, Petrek M (2018) Whole-exome sequencing identifies rare genetic variations in German families with pulmonary sarcoidosis. Hum Genet 137(9):705–716. https://doi.org/10.1007/s00439-018-1915-y
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Funding
Matthijs Brouwer is supported by a grant from the Netherlands Organization for Health Research and Development (ZonMw; NWO-Vidi grant 2019 [917.17.308]). Diederik van de Beek is supported by grants from the Netherlands Organization for Health Research and Development (ZonMw; NWO-Vici grant 2019 [918.19.627]), the European Research Council (ERC Starting Grant 281156), and an innovation grant by the board of directors of the Academic Medical Center, Amsterdam, The Netherlands. No potential conflict of interest relevant for this article exists.
Author information
Authors and Affiliations
Contributions
DF and BF had a substantial contribution to conception and design, acquisition of data, analysis, and interpretation of data; drafted the manuscript; and final approval of the version to be published. MB had a substantial contribution to conception and design, acquisition of data, analysis, and interpretation of data; drafted the manuscript; and final approval of the version to be published. DvB had a substantial contribution to conception and design, acquisition of data, analysis, and interpretation of data; revised the manuscript for important intellectual content; and final approval of the version to be published.
Corresponding author
Ethics declarations
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary figure 1.
Flow chart of variant selection (DOCX 168 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fritz, D., Ferwerda, B., Brouwer, M.C. et al. Whole genome sequencing identifies variants associated with sarcoidosis in a family with a high prevalence of sarcoidosis. Clin Rheumatol 40, 3735–3743 (2021). https://doi.org/10.1007/s10067-021-05684-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05684-w