Abstract
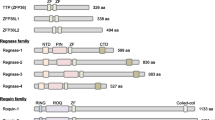
The transcription factors Bach1 and Bach2, which belong to a basic region-leucine zipper (bZip) family, repress target gene expression by forming heterodimers with small Maf proteins. With the ability to bind to heme, Bach1 and Bach2 are important in maintaining heme homeostasis in response to oxidative stress, which is characterized by high levels of reactive oxygen species (ROS) in cells and thereby induces cellular damage and senescence. The inactivation of Bach1 exerts an antioxidant effect. Thus, Bach1 may be a potential therapeutic target of oxidative stress-related diseases. Bach2 participates in oxidative stress-mediated apoptosis and is involved in macrophage-mediated innate immunity as well as the adaptive immune response. Bach1 and Bach2 promote the differentiation of common lymphoid progenitors to B cells by repressing myeloid-related genes. Bach2 is able to regulate class-switch recombination and plasma cell differentiation by altering the concentration of mitochondrial ROS during B cell differentiation. Furthermore, Bach2 maintains T cell homeostasis, influences the function of macrophages, and plays a role in autoimmunity. Bach2-controlling genes with super enhancers in T cells play a key role in immune regulation. However, in spite of new research, the role of Bach1 and Bach2 in immune cells and immune response is not completely clear, nor are their respective roles of in oxidative stress and the immune response, in particular with regard to the clinical phenotypes of autoimmune diseases. The anti-immunosenescence action of Bach and the role of epigenetic modifications of these transcription factors may be important in the mechanism of Bach transcription factors in mediating oxidative stress and cellular immunity.




Similar content being viewed by others
References
Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M et al (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Molecular and cellular biology 16:6083–95
Amoutzias GD, Veron AS, Weiner J 3rd, Robinson-Rechavi M, Bornberg-Bauer E, Oliver SG et al (2007) One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Molecular biology and evolution 24:827–35
Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D (1995) The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research 6:1193–8
Igarashi K, Watanabe-Matsui M (2014) Wearing red for signaling: the heme-Bach axis in heme metabolism, oxidative stress response and iron immunology. The Tohoku journal of experimental medicine 232:229–53
Rosbrook GO, Stead MA, Carr SB, Wright SC (2012) The structure of the Bach2 POZ-domain dimer reveals an intersubunit disulfide bond. Acta crystallographica Section D, Biological crystallography 68:26–34
Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S et al (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. The EMBO journal 20:2835–43
Watanabe-Matsui M, Muto A, Matsui T, Itoh-Nakadai A, Nakajima O, Murayama K et al (2011) Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood 117:5438–48
Watanabe-Matsui M, Matsumoto T, Matsui T, Ikeda-Saito M, Muto A, Murayama K et al (2015) Heme binds to an intrinsically disordered region of Bach2 and alters its conformation. Archives of biochemistry and biophysics 565:25–31
Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H et al (1998) Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. The Journal of biological chemistry 273:11783–90
Hoshino H, Igarashi K (2002) Expression of the oxidative stress-regulated transcription factor bach2 in differentiating neuronal cells. Journal of biochemistry 132:427–31
Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH (1993) Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722–8
Miller DM, Wang JA, Buchanan AK, Hall ED (2014) Temporal and spatial dynamics of nrf2-antioxidant response elements mediated gene targets in cortex and hippocampus after controlled cortical impact traumatic brain injury in mice. Journal of neurotrauma 31:1194–201
Taguchi K, Motohashi H, Yamamoto M (2011) Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes to cells : devoted to molecular & cellular mechanisms 16:123–40
Jyrkkanen HK, Kuosmanen S, Heinaniemi M, Laitinen H, Kansanen E, Mella-Aho E et al (2011) Novel insights into the regulation of antioxidant-response-element-mediated gene expression by electrophiles: induction of the transcriptional repressor BACH1 by Nrf2. The Biochemical journal 440:167–74
Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H et al (2000) Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. The Journal of biological chemistry 275:15370–6
Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y et al (2004) Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. The EMBO journal 23:2544–53
Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K (2003) Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. The Journal of biological chemistry 278:49246–53
Igarashi K, Ochiai K, Itoh-Nakadai A, Muto A (2014) Orchestration of plasma cell differentiation by Bach2 and its gene regulatory network. Immunological reviews 261:116–25
Ryter SW, Tyrrell RM (2000) The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free radical biology & medicine 28:289–309
Shibahara S (2003) The heme oxygenase dilemma in cellular homeostasis: new insights for the feedback regulation of heme catabolism. The Tohoku journal of experimental medicine 200:167–86
Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K (2004) Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proceedings of the National Academy of Sciences of the United States of America 101:1461–6
Tanimura N, Miller E, Igarashi K, Yang D, Burstyn JN, Dewey CN et al (2016) Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO reports 17:249–65
Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H et al (2002) Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. The EMBO journal 21:5216–24
Furuyama K, Kaneko K, Vargas PD (2007) Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. The Tohoku journal of experimental medicine 213:1–16
Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, Furuyama K et al (2003) Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. The Journal of biological chemistry 278:9125–33
Hira S, Tomita T, Matsui T, Igarashi K, Ikeda-Saito M (2007) Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB life 59:542–51
Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T et al (2007) Heme induces ubiquitination and degradation of the transcription factor Bach1. Molecular and cellular biology 27:6962–71
Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T et al (2011) The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. The Journal of biological chemistry 286:23521–32
Watari Y, Yamamoto Y, Brydun A, Ishida T, Mito S, Yoshizumi M et al (2008) Ablation of the bach1 gene leads to the suppression of atherosclerosis in bach1 and apolipoprotein E double knockout mice. Hypertension research : official journal of the Japanese Society of Hypertension 31:783–92
Chapple SJ, Keeley TP, Mastronicola D, Arno M, Vizcay-Barrena G, Fleck R et al (2015) Bach1 differentially regulates distinct Nrf2-dependent genes in human venous and coronary artery endothelial cells adapted to physiological oxygen levels. Free radical biology & medicine 92:152–162
Mito S, Ozono R, Oshima T, Yano Y, Watari Y, Yamamoto Y et al (2008) Myocardial protection against pressure overload in mice lacking Bach1, a transcriptional repressor of heme oxygenase-1. Hypertension 51:1570–7
Tanimoto T, Hattori N, Senoo T, Furonaka M, Ishikawa N, Fujitaka K et al (2009) Genetic ablation of the Bach1 gene reduces hyperoxic lung injury in mice: role of IL-6. Free radical biology & medicine 46:1119–26
Harusato A, Naito Y, Takagi T, Uchiyama K, Mizushima K, Hirai Y et al (2011) Suppression of indomethacin-induced apoptosis in the small intestine due to Bach1 deficiency. Free radical research 45:717–27
Inoue M, Tazuma S, Kanno K, Hyogo H, Igarashi K, Chayama K (2011) Bach1 gene ablation reduces steatohepatitis in mouse MCD diet model. Journal of clinical biochemistry and nutrition 48:161–6
Kondo K, Ishigaki Y, Gao J, Yamada T, Imai J, Sawada S et al (2013) Bach1 deficiency protects pancreatic beta-cells from oxidative stress injury. American journal of physiology Endocrinology and metabolism 305:E641–8
So AY, Garcia-Flores Y, Minisandram A, Martin A, Taganov K, Boldin M et al (2012) Regulation of APC development, immune response, and autoimmunity by Bach1/HO-1 pathway in mice. Blood 120:2428–37
Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S et al (2012) Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology 13:1118–28
Hama M, Kirino Y, Takeno M, Takase K, Miyazaki T, Yoshimi R et al (2012) Bach1 regulates osteoclastogenesis in a mouse model via both heme oxygenase 1-dependent and heme oxygenase 1-independent pathways. Arthritis and rheumatism 64:1518–28
Takada T, Miyaki S, Ishitobi H, Hirai Y, Nakasa T, Igarashi K et al (2015) Bach1 deficiency reduces severity of osteoarthritis through upregulation of heme oxygenase-1. Arthritis research & therapy 17:285
Muto A, Tashiro S, Tsuchiya H, Kume A, Kanno M, Ito E et al (2002) Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. The Journal of biological chemistry 277:20724–33
Yoshida C, Yoshida F, Sears DE, Hart SM, Ikebe D, Muto A et al (2007) Bcr-Abl signaling through the PI-3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase-1. Blood 109:1211–9
Casolari DA, Makri M, Yoshida C, Muto A, Igarashi K, Melo JV (2013) Transcriptional suppression of BACH2 by the Bcr-Abl oncoprotein is mediated by PAX5. Leukemia 27:409–15
Chen Z, Pittman EF, Romaguera J, Fayad L, Wang M, Neelapu SS et al (2013) Nuclear translocation of B-cell-specific transcription factor, BACH2, modulates ROS mediated cytotoxic responses in mantle cell lymphoma. PloS one 8:e69126
Ando R, Shima H, Tamahara T, Sato Y, Watanabe-Matsui M, Kato H et al (2016) The transcription factor Bach2 is phosphorylated at multiple sites in murine B cells but a single site prevents its nuclear localization. J Biol Chem 291:1826–40
Tashiro S, Muto A, Tanimoto K, Tsuchiya H, Suzuki H, Hoshino H et al (2004) Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Molecular and cellular biology 24:3473–84
Kono K, Harano Y, Hoshino H, Kobayashi M, Bazett-Jones DP, Muto A et al (2008) The mobility of Bach2 nuclear foci is regulated by SUMO-1 modification. Experimental cell research 314:903–13
Hoshino H, Nishino TG, Tashiro S, Miyazaki M, Ohmiya Y, Igarashi K et al (2007) Co-repressor SMRT and class II histone deacetylases promote Bach2 nuclear retention and formation of nuclear foci that are responsible for local transcriptional repression. Journal of biochemistry 141:719–27
Hong SW, Kim S, Lee DK (2008) The role of Bach2 in nucleic acid-triggered antiviral innate immune responses. Biochemical and biophysical research communications 365:426–32
Martensson IL, Almqvist N, Grimsholm O, Bernardi AI (2010) The pre-B cell receptor checkpoint. FEBS letters 584:2572–9
Melchers F (2005) The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nature reviews Immunology 5:578–84
Zouali M (2014) Transcriptional and metabolic pre-B cell receptor-mediated checkpoints: implications for autoimmune diseases. Molecular immunology 62:315–20
Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H et al (2011) Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood 118:4174–8
Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H et al (2013) BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nature medicine 19:1014–22
Swaminathan S, Duy C, Muschen M (2014) BACH2-BCL6 balance regulates selection at the pre-B cell receptor checkpoint. Trends in immunology 35:131–7
McAllister K, Yarwood A, Bowes J, Orozco G, Viatte S, Diogo D et al (2013) Identification of BACH2 and RAD51B as rheumatoid arthritis susceptibility loci in a meta-analysis of genome-wide data. Arthritis and rheumatism 65:3058–62
Hendriks RW, Middendorp S (2004) The pre-BCR checkpoint as a cell-autonomous proliferation switch. Trends in immunology 25:249–56
Oracki SA, Walker JA, Hibbs ML, Corcoran LM, Tarlinton DM (2010) Plasma cell development and survival. Immunological reviews 237:140–59
Shlomchik MJ, Weisel F (2012) Germinal center selection and the development of memory B and plasma cells. Immunological reviews 247:52–63
Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD (2011) The genetic network controlling plasma cell differentiation. Seminars in immunology 23:341–9
Dave SS (2014) The polyphony of BACH2. Blood 123:950
Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E et al (2004) The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429:566–71
Mendez A, Mendoza L (2016) A network model to describe the terminal differentiation of B cells. PLoS computational biology 12:e1004696
Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H et al (2006) Plasmacytic transcription factor blimp-1 is repressed by Bach2 in B cells. The Journal of biological chemistry 281:38226–34
Nera KP, Kohonen P, Narvi E, Peippo A, Mustonen L, Terho P et al (2006) Loss of Pax5 promotes plasma cell differentiation. Immunity 24:283–93
Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K (2008) Regulation of the plasma cell transcription factor blimp-1 gene by Bach2 and Bcl6. International immunology 20:453–60
Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A et al (2002) Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17:51–62
Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H (2006) Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25:225–36
Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R et al (2013) Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38:918–29
Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T et al (2006) Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nature immunology 7:773–82
Muto A, Ochiai K, Kimura Y, Itoh-Nakadai A, Calame KL, Ikebe D et al (2010) Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. The EMBO journal 29:4048–61
Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S et al (2013) Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity 39:136–47
Tanaka H, Muto A, Shima H, Katoh Y, Sax N, Tajima S et al. (2016) Epigenetic regulation of the Blimp-1 gene in B cells involves Bach2 and histone deacetylase 3
Itoh-Nakadai A, Hikota R, Muto A, Kometani K, Watanabe-Matsui M, Sato Y et al (2014) The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nature immunology 15:1171–80
Igarashi K, Itoh-Nakadai A (2016) Orchestration of B lymphoid cells and their inner myeloid by Bach. Current opinion in immunology 39:136–42
Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C et al (2009) Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457:318–21
Xie H, Ye M, Feng R, Graf T (2004) Stepwise reprogramming of B cells into macrophages. Cell 117:663–76
Jang KJ, Mano H, Aoki K, Hayashi T, Muto A, Nambu Y et al (2015) Mitochondrial function provides instructive signals for activation-induced B-cell fates. Nature communications 6:6750
Schumacher A, Zenclussen AC (2014) Effects of heme oxygenase-1 on innate and adaptive immune responses promoting pregnancy success and allograft tolerance. Frontiers in pharmacology 5:288
Kurotaki D, Uede T, Tamura T (2015) Functions and development of red pulp macrophages. Microbiology and immunology 59:55–62
Nakamura A, Ebina-Shibuya R, Itoh-Nakadai A, Muto A, Shima H, Saigusa D et al (2013) Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. The Journal of experimental medicine 210:2191–204
Tsukumo S, Unno M, Muto A, Takeuchi A, Kometani K, Kurosaki T et al (2013) Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proceedings of the National Academy of Sciences of the United States of America 110:10735–40
Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M et al (2013) BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498:506–10
Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M (2014) Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. Journal of immunology 192:985–95
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–19
Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR et al (2015) Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520:558–62
Hu G, Chen J (2013) A genome-wide regulatory network identifies key transcription factors for memory CD8(+) T-cell development. Nature communications 4:2830
Chang C (2014) Autoimmunity: from black water fever to regulatory function. J Autoimmun 48–49:1–9
Ghodke-Puranik Y, Niewold TB (2015) Immunogenetics of systemic lupus erythematosus: a comprehensive review. J Autoimmun 64:125–36
Kuhn A, Landmann A (2014) The classification and diagnosis of cutaneous lupus erythematosus. J Autoimmun 48–49:14–9
Kourilovitch M, Galarza-Maldonado C, Ortiz-Prado E (2014) Diagnosis and classification of rheumatoid arthritis. J Autoimmun 48–49:26–30
Eisenstein EM, Berkun Y (2014) Diagnosis and classification of juvenile idiopathic arthritis. J Autoimmun 48–49:31–3
Alunno A, Carubbi F, Bistoni O, Caterbi S, Bartoloni E, Bigerna B et al (2014) CD4(−)CD8(−) T-cells in primary Sjogren’s syndrome: association with the extent of glandular involvement. J Autoimmun 51:38–43
Colafrancesco S, Perricone C, Priori R, Valesini G, Shoenfeld Y (2014) Sjogren’s syndrome: another facet of the autoimmune/inflammatory syndrome induced by adjuvants (ASIA). J Autoimmun 51:10–6
Goules AV, Tzioufas AG, Moutsopoulos HM (2014) Classification criteria of Sjogren’s syndrome. J Autoimmun 48–49:42–5
Moutsopoulos HM (2014) Sjogren’s syndrome: a forty-year scientific journey. J Autoimmun 51:1–9
Harden JL, Krueger JG, Bowcock AM (2015) The immunogenetics of psoriasis: a comprehensive review. J Autoimmun 64:66–73
Kallenberg CG (2014) The diagnosis and classification of microscopic polyangiitis. J Autoimmun 48–49:90–3
Hernandez-Rodriguez J, Alba MA, Prieto-Gonzalez S, Cid MC (2014) Diagnosis and classification of polyarteritis nodosa. J Autoimmun 48–49:84–9
Baughman RP, Lower EE (2015) Treatment of sarcoidosis. Clin Rev Allergy Immunol 49:79–92
Wessendorf TE, Bonella F, Costabel U (2015) Diagnosis of sarcoidosis. Clin Rev Allergy Immunol 49:54–62
Chen ES, Moller DR (2015) Etiologies of sarcoidosis. Clin Rev Allergy Immunol 49:6–18
Rossi G, Cavazza A, Colby TV (2015) Pathology of sarcoidosis. Clin Rev Allergy Immunol 49:36–44
Spagnolo P (2015) Sarcoidosis: a critical review of history and milestones. Clin Rev Allergy Immunol 49:1–5
Hollenbach JA, Oksenberg JR (2015) The immunogenetics of multiple sclerosis: a comprehensive review. J Autoimmun 64:13–25
Bowlus CL, Gershwin ME (2014) The diagnosis of primary biliary cirrhosis. Autoimmun Rev 13:441–4
Efe C, Kahramanoglu-Aksoy E, Yilmaz B, Ozseker B, Takci S, Roach EC et al (2014) Pregnancy in women with primary biliary cirrhosis. Autoimmun Rev 13:931–5
Floreani A, Infantolino C, Franceschet I, Tene IM, Cazzagon N, Buja A et al (2015) Pregnancy and primary biliary cirrhosis: a case–control study. Clin Rev Allergy Immunol 48:236–42
Sun Y, Haapanen K, Li B, Zhang W, Van de Water J, Gershwin ME (2015) Women and primary biliary cirrhosis. Clin Rev Allergy Immunol 48:285–300
Webb GJ, Siminovitch KA, Hirschfield GM (2015) The immunogenetics of primary biliary cirrhosis: a comprehensive review. J Autoimmun 64:42–52
Lee HJ, Li CW, Hammerstad SS, Stefan M, Tomer Y (2015) Immunogenetics of autoimmune thyroid diseases: a comprehensive review. J Autoimmun 64:82–90
Bossini-Castillo L, Lopez-Isac E, Martin J (2015) Immunogenetics of systemic sclerosis: defining heritability, functional variants and shared-autoimmunity pathways. J Autoimmun 64:53–65
Hudson M, Fritzler MJ (2014) Diagnostic criteria of systemic sclerosis. J Autoimmun 48–49:38–41
Wang Q, Yang F, Miao Q, Krawitt EL, Gershwin ME, Ma X (2016) The clinical phenotypes of autoimmune hepatitis: a comprehensive review. J Autoimmun 66:98–107
Aricha R, Reuveni D, Fuchs S, Souroujon MC (2016) Suppression of experimental autoimmune myasthenia gravis by autologous T regulatory cells. J Autoimmun 67:57–64
Avidan N, Le Panse R, Berrih-Aknin S, Miller A (2014) Genetic basis of myasthenia gravis—a comprehensive review. J Autoimmun 52:146–53
Berrih-Aknin S (2014) Myasthenia gravis: paradox versus paradigm in autoimmunity. J Autoimmun 52:1–28
Berrih-Aknin S, Le Panse R (2014) Myasthenia gravis: a comprehensive review of immune dysregulation and etiological mechanisms. J Autoimmun 52:90–100
Canivell S, Gomis R (2014) Diagnosis and classification of autoimmune diabetes mellitus. Autoimmun Rev 13:403–7
Ferretti C, La Cava A (2016) Adaptive immune regulation in autoimmune diabetes. Autoimmun Rev 15:236–41
Tan T, Xiang Y, Chang C, Zhou Z (2014) Alteration of regulatory T cells in type 1 diabetes mellitus: a comprehensive review. Clin Rev Allergy Immunol 47:234–43
Xie Z, Chang C, Zhou Z (2014) Molecular mechanisms in autoimmune type 1 diabetes: a critical review. Clin Rev Allergy Immunol 47:174–92
Noble JA (2015) Immunogenetics of type 1 diabetes: a comprehensive review. J Autoimmun 64:101–12
Perga S, Montarolo F, Martire S, Berchialla P, Malucchi S, Bertolotto A (2015) Anti-inflammatory genes associated with multiple sclerosis: a gene expression study. Journal of neuroimmunology 279:75–8
Hoppmann N, Graetz C, Paterka M, Poisa-Beiro L, Larochelle C, Hasan M et al (2015) New candidates for CD4 T cell pathogenicity in experimental neuroinflammation and multiple sclerosis. Brain : a journal of neurology 138:902–17
Kiani AK, Jahngir S, John P, Bhatti A, Zia A, Wang X et al (2015) Genetic link of type 1 diabetes susceptibility loci with rheumatoid arthritis in Pakistani patients. Immunogenetics 67:277–82
Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P et al (2011) Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378:1006–14
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T et al (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature genetics 42:1118–25
Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T et al (2011) Abundant pleiotropy in human complex diseases and traits. American journal of human genetics 89:607–18
Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A et al (2010) Multiple common variants for celiac disease influencing immune gene expression. Nature genetics 42:295–302
Quinn EM, Coleman C, Molloy B, Dominguez Castro P, Cormican P, Trimble V et al (2015) Transcriptome analysis of CD4+ T cells in coeliac disease reveals imprint of BACH2 and IFNgamma regulation. PloS one 10:e0140049
Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL et al (2012) Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nature genetics 44:676–80
Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE et al (2008) Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nature genetics 40:1399–401
Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C et al (2011) Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS genetics 7:e1002216
Elboudwarej E, Cole M, Briggs FB, Fouts A, Fain PR, Quach H et al (2016) Hypomethylation within gene promoter regions and type 1 diabetes in discordant monozygotic twins. Journal of autoimmunity 68:23–9
Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM (2011) Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome research 21:1109–21
Christodoulou K, Wiskin AE, Gibson J, Tapper W, Willis C, Afzal NA et al (2013) Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut 62:977–84
Medici M, Porcu E, Pistis G, Teumer A, Brown SJ, Jensen RA et al (2014) Identification of novel genetic loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS genetics 10:e1004123
Graham DB, Xavier RJ (2013) From genetics of inflammatory bowel disease towards mechanistic insights. Trends in immunology 34:371–8
Parkes M, Cortes A, van Heel DA, Brown MA (2013) Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nature reviews Genetics 14:661–73
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–33
Lu Q (2013) The critical importance of epigenetics in autoimmunity. Journal of autoimmunity 41:1–5
Saito Y, Saito H, Liang G, Friedman JM (2014) Epigenetic alterations and microRNA misexpression in cancer and autoimmune diseases: a critical review. Clin Rev Allergy Immunol 47:128–35
Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT et al (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–9
Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC et al (2007) Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. Journal of virology 81:12836–45
Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR et al (2007) Requirement of bic/microRNA-155 for normal immune function. Science (New York, NY) 316:608–11
Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y et al (2007) Regulation of the germinal center response by microRNA-155. Science (New York, NY) 316:604–8
O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D (2007) MicroRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences of the United States of America 104:1604–9
Belver L, Papavasiliou FN, Ramiro AR (2011) MicroRNA control of lymphocyte differentiation and function. Current opinion in immunology 23:368–73
Fernando TR, Rodriguez-Malave NI, Rao DS (2012) MicroRNAs in B cell development and malignancy. Journal of hematology & oncology 5:7
Porstner M, Winkelmann R, Daum P, Schmid J, Pracht K, Corte-Real J et al (2015) miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors mitf and Bach2. European journal of immunology 45:1206–15
Haam K, Kim HJ, Lee KT, Kim JH, Kim M, Kim SY et al (2014) Epigenetic silencing of BTB and CNC homology 2 and concerted promoter CpG methylation in gastric cancer. Cancer letters 351:206–14
Igarashi K, Ota K, Nakame A (2009) Regulation of cellular senescence by Bach1. Nihon rinsho Japanese journal of clinical medicine 67:1423–8
Omura S, Suzuki H, Toyofuku M, Ozono R, Kohno N, Igarashi K (2005) Effects of genetic ablation of bach1 upon smooth muscle cell proliferation and atherosclerosis after cuff injury. Genes to cells : devoted to molecular & cellular mechanisms 10:277–85
Kuwahara M, Suzuki J, Tofukuji S, Yamada T, Kanoh M, Matsumoto A et al (2014) The menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nature communications 5:3555
Montoya-Ortiz G (2013) Immunosenescence, aging, and systemic lupus erythematous. Autoimmune diseases 2013:267078
Vieira SA, Deininger MW, Sorour A, Sinclair P, Foroni L, Goldman JM et al (2001) Transcription factor BACH2 is transcriptionally regulated by the BCR/ABL oncogene. Genes, chromosomes & cancer 32:353–63
Ichikawa S, Fukuhara N, Katsushima H, Takahashi T, Yamamoto J, Yokoyama H et al (2014) Association between BACH2 expression and clinical prognosis in diffuse large B-cell lymphoma. Cancer science 105:437–44
Roychoudhuri R, Eil RL, Clever D, Klebanoff CA, Sukumar M, Grant FM et al (2016) The transcription factor BACH2 promotes tumor immunosuppression. The Journal of clinical investigation 126:599–604
Acknowledgments
This work is supported by grants from the National Natural Science Foundation of China (No. 81220108017, No. 81430074, No. 81270024, and No. 30972745).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, Y., Wu, H., Zhao, M. et al. The Bach Family of Transcription Factors: A Comprehensive Review. Clinic Rev Allerg Immunol 50, 345–356 (2016). https://doi.org/10.1007/s12016-016-8538-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-016-8538-7