Abstract
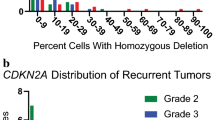
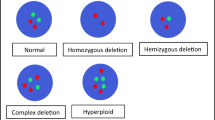
Deletion of CDKN2A occurs in 50% of glioblastomas (GBM), and IFNA locus deletion in 25%. These genes reside closely on chromosome 9. We investigated whether CDKN2A and IFNA were co-deleted within the same heterogeneous tumour and their prognostic implications. We assessed CDKN2A and IFNA14 deletions in 45 glioma samples using an in-house three-colour FISH probe. We examined the correlation between p16INK4a protein expression (via IHC) and CDKN2A deletion along with the impact of these genomic events on patient survival. FISH analyses demonstrated that grades II and III had either wildtype (wt) or amplified CDKN2A/IFNA14, whilst 44% of GBMs harboured homozygous deletions of both genes. Cores with CDKN2A homozygous deletion (n = 11) were negative for p16INK4a. Twenty p16INK4a positive samples lacked CDKN2A deletion with some of cells showing negative p16INK4a. There was heterogeneity in IFNA14/CDKN2A ploidy within each GBM. Survival analyses of primary GBMs suggested a positive association between increased p16INK4a and longer survival; this persisted when considering CDKN2A/IFNA14 status. Furthermore, wt (intact) CDKN2A/IFNA14 were found to be associated with longer survival in recurrent GBMs. Our data suggest that co-deletion of CDKN2A/IFNA14 in GBM negatively correlates with survival and CDKN2A-wt status correlated with longer survival, and with second surgery, itself a marker for improved patient outcomes.






Similar content being viewed by others
Data availability
The patients’ datasets used in this work are not publicly available due to patients’ privacy concerns of the institutional review board policies on human tissue data. The CDKN2A and IFNs gene status profiles that were utilised in this work are available from the cBioPortal database.
References
Weller M et al (2015) Glioma. Nat Rev Dis Prim 1:15017
Ostrom QT et al (2019) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol 21(Suppl 5):v1–v100
Stupp R et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996
Stupp R et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466
Hanif F et al (2017) Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev 18(1):3–9
Verhaak RG et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110
Brennan CW et al (2013) The somatic genomic landscape of glioblastoma. Cell 155(2):462–477
Cancer Genome Atlas Research (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068
Louis DN et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Louis DN et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251
Gonzalez-Castro LN, Wesseling P (2021) The cIMPACT-NOW updates and their significance to current neuro-oncology practice. Neurooncol Pract 8(1):4–10
Ma S et al (2020) Prognostic impact of CDKN2A/B deletion, TERT mutation, and EGFR amplification on histological and molecular IDH-wildtype glioblastoma. Neurooncol Adv 2(1):vdaa126
Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7(9):667–677
Kim WY, Sharpless NE (2006) The regulation of INK4/ARF in cancer and aging. Cell 127(2):265–275
Lu VM et al (2020) The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol 148(2):221–229
Park JW et al (2021) The prognostic significance of p16 expression pattern in diffuse gliomas. J Pathol Transl Med 55(2):102–111
Appay R et al (2019) CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol 21(12):1519–1528
Reis GF et al (2015) CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol 74(5):442–452
Romagosa C et al (2011) p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30(18):2087–2097
Milde-Langosch K et al (2001) Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat 67(1):61–70
Lee CT et al (1999) Overexpression of the cyclin-dependent kinase inhibitor p16 is associated with tumor recurrence in human prostate cancer. Clin Cancer Res 5(5):977–983
Gutiontov SI et al (2021) CDKN2A loss-of-function predicts immunotherapy resistance in non-small cell lung cancer. Sci Rep 11(1):20059
Parkin J, Cohen B (2001) An overview of the immune system. Lancet 357(9270):1777–1789
de Padilla CML, Niewold TB (2016) The type I interferons: basic concepts and clinical relevance in immune-mediated inflammatory diseases. Gene 576(1):14–21
Ferrantini M, Capone I, Belardelli F (2007) Interferon-alpha and cancer: mechanisms of action and new perspectives of clinical use. Biochimie 89(6–7):884–893
Vidal P (2020) Interferon α in cancer immunoediting: from elimination to escape. Scand J Immunol 91(5):e12863
Tarhini AA, Gogas H, Kirkwood JM (2012) IFN-α in the treatment of melanoma. J Immunol 189(8):3789–3793
Kankuri-Tammilehto M et al (2023) Long-term outcome with prolonged use of interferon-alpha administered intermittently for metastatic renal cell carcinoma: a phase II study. Anticancer Res 43(6):2645–2657
Guo J et al (2019) Empowering therapeutic antibodies with IFN-α for cancer immunotherapy. PLoS ONE 14(8):e0219829
Guo C et al (2023) Adjuvant temozolomide chemotherapy with or without interferon Alfa among patients with newly diagnosed high-grade gliomas: a randomized clinical trial. JAMA Netw Open 6(1):e2253285
Fujita M et al (2010) Role of type 1 IFNs in antiglioma immunosurveillance–using mouse studies to guide examination of novel prognostic markers in humans. Clin Cancer Res 16(13):3409–3419
Yu R, Zhu B, Chen D (2022) Type I interferon-mediated tumor immunity and its role in immunotherapy. Cell Mol Life Sci 79(3):191
Rafique I, Kirkwood JM, Tarhini AA (2015) Immune checkpoint blockade and interferon-α in melanoma. Semin Oncol 42(3):436–447
Aricò E et al (2019) Type I interferons and cancer: an evolving story demanding novel clinical applications. Cancers (Basel) 11(12):1943
Tarhini AA et al (2012) Differing patterns of circulating regulatory T cells and myeloid-derived suppressor cells in metastatic melanoma patients receiving anti-CTLA4 antibody and interferon-α or TLR-9 agonist and GM-CSF with peptide vaccination. J Immunother 35(9):702–710
Al Shboul S et al (2021) Kinomics platform using GBM tissue identifies BTK as being associated with higher patient survival. Life Sci Alliance 4(12):e202101054
Kononen J et al (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4(7):844–847
Purkait S et al (2013) CDKN2A deletion in pediatric versus adult glioblastomas and predictive value of p16 immunohistochemistry. Neuropathology 33(4):405–412
Jubb A, Boyle S (2020) Visualizing genome reorganization using 3D DNA FISH. Methods Mol Biol 2148:85–95
Boyle S et al (2011) Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res 19(7):901–909
Boyle S et al (2020) A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev 34(13–14):931–949
Pollard SM et al (2009) Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4(6):568–580
Schindelin J et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Bankhead P et al (2017) QuPath: open source software for digital pathology image analysis. Sci Rep 7(1):16878
Delaunay T et al (2020) Frequent homozygous deletions of type I interferon genes in pleural mesothelioma confer sensitivity to oncolytic measles virus. J Thorac Oncol 15(5):827–842
Kim UJ et al (1992) Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res 20(5):1083–1085
Brennan PM et al (2021) Second surgery for progressive glioblastoma: a multi-centre questionnaire and cohort-based review of clinical decision-making and patient outcomes in current practice. J Neurooncol 153(1):99–107
Cottone L et al (2020) Frequent alterations in p16/CDKN2A identified by immunohistochemistry and FISH in chordoma. J Pathol Clin Res 6(2):113–123
Kamiryo T et al (2002) Analysis of homozygous deletion of the p16 gene and correlation with survival in patients with glioblastoma multiforme. J Neurosurg 96(5):815–822
Burns KL et al (1998) Molecular genetic correlates of p16, cdk4, and pRb immunohistochemistry in glioblastomas. J Neuropathol Exp Neurol 57(2):122–130
Wang H et al (2020) Identification of genomic alterations and associated transcriptomic profiling reveal the prognostic significance of MMP14 and PKM2 in patients with pancreatic cancer. Aging (Albany NY) 12(18):18676–18692
Worst TS et al (2018) CDKN2A as transcriptomic marker for muscle-invasive bladder cancer risk stratification and therapy decision-making. Sci Rep 8(1):14383
Chen Z et al (2021) Comprehensive analysis revealed that CDKN2A is a biomarker for immune infiltrates in multiple cancers. Front Cell Dev Biol 9:808208
Liu W et al (2020) Loss of CDKN2A at chromosome 9 has a poor clinical prognosis and promotes lung cancer progression. Mol Genet Genom Med 8(12):e1521
Peng Y et al (2022) Co-occurrence of CDKN2A/B and IFN-I homozygous deletions correlates with an immunosuppressive phenotype and poor prognosis in lung adenocarcinoma. Mol Oncol 16(8):1746–1760
Ye Z et al (2018) Prevalent homozygous deletions of type I interferon and defensin genes in human cancers associate with immunotherapy resistance. Clin Cancer Res 24(14):3299–3308
Barriga FM et al (2022) MACHETE identifies interferon-encompassing chromosome 9p21.3 deletions as mediators of immune evasion and metastasis. Nature Cancer 3(11):1367–1385
Nassar A et al (2010) Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Appl Immunohistochem Mol Morphol 18(5):433–441
Kündig P et al (2018) Limited utility of tissue micro-arrays in detecting intra-tumoral heterogeneity in stem cell characteristics and tumor progression markers in breast cancer. J Transl Med 16(1):118
Acknowledgements
We would like to thank Helen Caldwell from the pathology department at the University of Edinburgh for IHC preparation and staining. We would like to thank the members of the Advanced Imaging Resource facility at the Institute of Genetics and Cancer (IGC) for facilitating the use of microscopes and image processing software. The authors are sincerely thankful to NHS Lothian Bioresource for granting access to tissue samples.
Funding
Sofian Al Shboul (SAS) and Tareq Saleh (TS) are supported by the Deanship of Scientific Research, The Hashemite University (SAS: grants no. 785/48/2022 and 738/54/2022; TS: grants no. 465/83/2019 and 418/84/2019).
Author information
Authors and Affiliations
Contributions
SAS: conceptualization, acquiring FFPE FISH and IHC images, analysis of FISH and IHC data, designing and creating the figures and tables, writing—original draft, review, and editing. SB: designed and labelled FISH probes, conducted FISH on the FFPE slides. AS: conceptualization, data analysis and writing original draft. TS: FISH and IHC analysis, designing figures, writing—original draft, review, and editing. MA, OAK and SAB: performed pathological assessment of the protein marker expression. AM and RD: FISH image analysis. SG: performed FISH on cells and obtained the images. KB: conceptualization and supervision of the project. TH: conceptualization, resources, supervision of the project, funding acquisition, analysis of FISH and IHC data, designing and creating the figures and tables, writing—original draft, review, and editing. PMB: conceptualization, resources, acquired the FFPE samples and supervised the construction of the TMA, supervision of the project, analysis of FISH and IHC data, designing and creating the figures and tables, writing—original draft, review, and editing. *All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors have no relevant financial or non-financial interests to disclose.
Ethical approval
GBM FFPE samples used to construct the TMA were obtained under ethical approvals from the regional ethics committee (LREC 115/ES/0094).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Al Shboul, S., Boyle, S., Singh, A. et al. FISH analysis reveals CDKN2A and IFNA14 co-deletion is heterogeneous and is a prominent feature of glioblastoma. Brain Tumor Pathol 41, 4–17 (2024). https://doi.org/10.1007/s10014-023-00473-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-023-00473-6