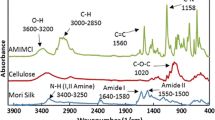
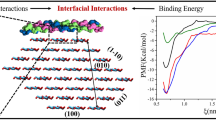
Abstract
Context
Due to their excellent biocompatibility and degradability, cellulose/spider silk protein composites hold a significant value in biomedical applications such as tissue engineering, drug delivery, and medical dressings. The interfacial interactions between cellulose and spider silk protein affect the properties of the composite. Therefore, it is important to understand the interfacial interactions between spider silk protein and cellulose to guide the design and optimization of composites. The study of the adsorption of protein on specific surfaces of cellulose crystal can be very complex using experimental methods. Molecular dynamics simulations allow the exploration of various physical and chemical changes at the atomic level of the material and enable an atomic description of the interactions between cellulose crystal planes and spider silk protein. In this study, molecular dynamics simulations were employed to investigate the interfacial interactions between spider silk protein (NTD) and cellulose surfaces. Findings of RMSD, RMSF, and secondary structure showed that the structure of NTD proteins remained unchanged during the adsorption process. Cellulose contact numbers and hydrogen bonding trends on different crystalline surfaces suggest that van der Waals forces and hydrogen bonding interactions drive the binding of proteins to cellulose. These findings reveal the interaction between cellulose and protein at the molecular level and provide theoretical guidance for the design and synthesis of cellulose/spider silk protein composites.
Methods
MD simulations were all performed using the GROMACS-5.1 software package and run with CHARMM36 carbohydrate force field. Molecular dynamics simulations were performed for 500 ns for the simulated system.












Similar content being viewed by others
Data availability
Data will be made available on request.
References
Bhat A, Khan I, Usmani MA, Umapathi R, Al-Kindy SM (2019) Cellulose an ageless renewable green nanomaterial for medical applications: an overview of ionic liquids in extraction, separation and dissolution of cellulose. Int J Biol Macromol 129:750–777. https://doi.org/10.1016/j.ijbiomac.2018.12.190
Römling U, Galperin MY (2015) Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol 23(9):545–557. https://doi.org/10.1016/j.tim.2015.05.005
Abdelhamid HN, Mathew AP (2021) Cellulose-based materials for water remediation: adsorption, catalysis, and antifouling. Front Chem Eng 3:790314. https://doi.org/10.3389/fceng.2021.790314
Akturk A (2023) Enrichment of cellulose acetate nanofibrous scaffolds with retinyl palmitate and clove essential oil for wound healing applications. ACS Omega 8(6):5553–5560. https://doi.org/10.1021/acsomega.2c06881
Mohamed MA, Abd Mutalib M, Hir ZAM, Zain M, Mohamad AB, Minggu LJ, Awang NA, Salleh W (2017) An overview on cellulose-based material in tailoring bio-hybrid nanostructured photocatalysts for water treatment and renewable energy applications. Int J Biol Macromol 103:1232–1256. https://doi.org/10.1016/j.ijbiomac.2017.05.181
Ghalei S, Douglass M, Handa H (2022) Nitric oxide-releasing nanofibrous scaffolds based on silk fibroin and zein with enhanced biodegradability and antibacterial properties. ACS Biomater Sci Eng 8(7):3066–3077. https://doi.org/10.1021/acsbiomaterials.2c00103
Rising A, Johansson J (2015) Toward spinning artificial spider silk. Nat Chem Biol 11(5):309–315. https://doi.org/10.1038/nchembio.1789
Vollrath F, Knight D (1999) Structure and function of the silk production pathway in the spider Nephila edulis. Int J Biol Macromol 24(2–3):243–249. https://doi.org/10.1016/S0141-8130(98)00095-6
Wendt H, Hillmer A, Reimers K, Kuhbier JW, Schäfer-Nolte F, Allmeling C, Kasper C, Vogt PM (2011) Artificial skin–culturing of different skin cell lines for generating an artificial skin substitute on cross-weaved spider silk fibres. PLoS ONE 6(7):e21833. https://doi.org/10.1371/journal.pone.0021833
Allmeling C, Jokuszies A, Reimers K, Kall S, Choi C, Brandes G, Kasper C, Scheper T, Guggenheim M, Vogt P (2008) Spider silk fibres in artificial nerve constructs promote peripheral nerve regeneration. Cell Prolif 41(3):408–420. https://doi.org/10.1111/j.1365-2184.2008.00534.x
Gellynck K, Verdonk PC, Van Nimmen E, Almqvist KF, Gheysens T, Schoukens G, Van Langenhove L, Kiekens P, Mertens J, Verbruggen G (2008) Silkworm and spider silk scaffolds for chondrocyte support. J Mater Sci Mater Med 19:3399–3409. https://doi.org/10.1007/s10856-008-3474-6
SM Choi, P Chaudhry, SM Zo and SS Han (2018) Advances in protein-based materials: from origin to novel biomaterials. Cutting-edge enabling technologies for regenerative medicine:161–210.https://doi.org/10.1007/978-981-13-0950-2_10
Lawrence BD, Marchant JK, Pindrus MA, Omenetto FG, Kaplan DL (2009) Silk film biomaterials for cornea tissue engineering. Biomaterials 30(7):1299–1308. https://doi.org/10.1002/macp.202170013
Kundu B, Rajkhowa R, Kundu SC, Wang X (2013) Silk fibroin biomaterials for tissue regenerations. Adv. Drug Delivery Rev. 65(4):457–470. https://doi.org/10.1016/j.addr.2012.09.043
Harris TI, Gaztambide DA, Day BA, Brock CL, Ruben AL, Jones JA, Lewis RV (2016) Sticky situation: an investigation of robust aqueous-based recombinant spider silk protein coatings and adhesives. Biomacromolecules 17(11):3761–3772. https://doi.org/10.1021/acs.biomac.6b01267
Lewicka M, Hermanson O, Rising AU (2012) Recombinant spider silk matrices for neural stem cell cultures. Biomaterials 33(31):7712–7717. https://doi.org/10.1016/j.biomaterials.2012.07.021
Franco AR, Fernandes EM, Rodrigues MT, Rodrigues FJ, Gomes ME, Leonor IB, Kaplan DL, Reis RL (2019) Antimicrobial coating of spider silk to prevent bacterial attachment on silk surgical sutures. Acta Biomater. 99:236–246. https://doi.org/10.1016/j.actbio.2019.09.004
Allardyce BJ, Rajkhowa R, Dilley RJ, Redmond SL, Atlas MD, Wang X (2017) Glycerol-plasticised silk membranes made using formic acid are ductile, transparent and degradation-resistant. Mater. Sci. Eng C 80:165–173. https://doi.org/10.1016/j.msec.2017.05.112
Mohammadi P, Beaune GG, Stokke BT, Timonen JV, Linder MB (2018) Self-coacervation of a silk-like protein and its use as an adhesive for cellulosic materials. ACS macro letters 7(9):1120–1125. https://doi.org/10.1021/acsmacrolett.8b00527
Mohammadi P, Aranko AS, Landowski CP, Ikkala O, Jaudzems K, Wagermaier W, Linder MB (2019) Biomimetic composites with enhanced toughening using silk-inspired triblock proteins and aligned nanocellulose reinforcements. Sci Adv 5(9):2541. https://doi.org/10.1126/sciadv.aaw2541
Meirovitch S, Shtein Z, Ben-Shalom T, Lapidot S, Tamburu C, Hu X, Kluge JA, Raviv U, Kaplan DL, Shoseyov O (2016) Spider silk-CBD-cellulose nanocrystal composites: mechanism of assembly. Int J Mol Sci 17(9):1573. https://doi.org/10.3390/ijms17091573
Lemetti L, Tersteegen J, Sammaljarvi J, Aranko AS, Linder MB, Engineering (2021) Recombinant spider silk protein and delignified wood form a strong adhesive system. ACS Sustain Chem Eng 10(1):552–561. https://doi.org/10.1021/acssuschemeng.1c07043
Voutilainen S, Paananen A, Lille M, Linder MB (2019) Modular protein architectures for pH-dependent interactions and switchable assembly of nanocellulose. Int J Biol Macromol 137:270–276. https://doi.org/10.1016/j.ijbiomac.2019.06.227
Jesson DA, Watts JF (2012) The interface and interphase in polymer matrix composites: effect on mechanical properties and methods for identification. Polym Rev (Philadelphia, PA, U.S) 52(3):321–354. https://doi.org/10.1080/15583724.2012.710288
Muthoka RM, Panicker PS, Kim J (2022) Molecular dynamics study of cellulose nanofiber alignment under an electric field. Polymers 14(9):1925. https://doi.org/10.3390/polym14091925
Atkison JH, Parnham S, Marcotte WR, Olsen SK (2016) Crystal structure of the nephila clavipes major ampullate spidroin 1A N-terminal domain reveals plasticity at the dimer interface. J. Biol. Chem. 291(36):19006–19017. https://doi.org/10.1074/jbc.M116.736710
Vermaas JV, Crowley MF, Beckham GT, Engineering (2019) A quantitative molecular atlas for interactions between lignin and cellulose. ACS Sustain Chem Eng 7(24):19570–19583. https://doi.org/10.1021/acssuschemeng.9b04648
Malaspina DC, Faraudo J (2019) Molecular insight into the wetting behavior and amphiphilic character of cellulose nanocrystals. Adv Colloid Interface Sci 267:15–25. https://doi.org/10.1016/j.cis.2019.02.003
Ren Z, Guo R, Bi H, Jia X, Xu M, Cai L, Interfaces (2019) Interfacial adhesion of polylactic acid on cellulose surface: a molecular dynamics study. ACS Appl Mater Interfaces 12(2):3236–3244. https://doi.org/10.1021/acsami.9b20101
Gomes TC, Skaf MS (2012) Cellulose-builder: a toolkit for building crystalline structures of cellulose. J Comput Chem 33(14):1338–1346. https://doi.org/10.1002/jcc.22959
Nishiyama Y, Sugiyama J, Chanzy H, Langan P (2003) Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 125(47):14300–14306. https://doi.org/10.1021/ja037055w
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124(31):9074–9082. https://doi.org/10.1021/ja0257319
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Boonstra S, Onck PR, van der Giessen E (2016) CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state. J Phys Chem B 120(15):3692–3698. https://doi.org/10.1021/acs.jpcb.6b01316
Liu Y, Song X, Yang Y (2020) Anisotropic protein diffusion on nanosurface. Nanoscale 12(8):5209–5216. https://doi.org/10.1039/c9nr08555f
Ge C, Du J, Zhao L, Wang L, Liu Y, Li D (2011) Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc Natl Acad Sci U.S.A 108(41):16968–16973. https://doi.org/10.1073/pnas.1105270108
Ma H, Shi Q, Li X, Ren J, Wang Y, Li Z, Ning L (2023) Molecular and thermodynamic insights into interfacial interactions between collagen and cellulose investigated by molecular dynamics simulation and umbrella sampling. J Comput-Aided Mol Des 37(1):39–51. https://doi.org/10.1007/s10822-022-00489-8
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Berendsen HJ, van der Spoel D, van Drunen R (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Lee S, Tran A, Allsopp M, Lim JB, Hénin J, Klauda JB (2014) CHARMM36 united atom chain model for lipids and surfactants. J Phys Chem B 118(2):547–556. https://doi.org/10.1021/jp410344g
French AD (2017) Glucose, not cellobiose, is the repeating unit of cellulose and why that is important. Cellulose 24:4605–4609. https://doi.org/10.1007/s10570-017-1450-3
Petersen HG (1995) Accuracy and efficiency of the particle mesh Ewald method. J Chem Phys 103(9):3668–3679. https://doi.org/10.1063/1.470043
Hess B, Bekker H, Berendsen HJ, Fraaije JG (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Berendsen HJ, Postma JV, Van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81(8):3684–3690. https://doi.org/10.1063/1.448118
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190. https://doi.org/10.1063/1.328693
Parrinello M, Rahman A (1998) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190. https://doi.org/10.1063/1.328693
Hub JS (2015) g_wham—a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Phys 6(9):3713–3720. https://doi.org/10.1063/1.4931819
Hooft RW, Sander C, Vriend G (1997) Objectively judging the quality of a protein structure from a Ramachandran plot. Comput Appl Biosci 13(4):425–430. https://doi.org/10.1093/bioinformatics/13.4.425
Sargsyan K, Grauffel C, Lim C (2017) How molecular size impacts RMSD applications in molecular dynamics simulations. J Chem Theory Comput. 13(4):1518–1524. https://doi.org/10.1021/acs.jctc.7b00028
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolym Orig Res Biomol 22(12):2577–2637. https://doi.org/10.1002/bip.360221211
Feng Q, Noumbissi R, Biswas K, Koizumi H (2017) The role of hydroxyl groups in interchain interactions in cellulose Iα and Iβ. Int J Quantum Chem 117(10):e25357. https://doi.org/10.1002/qua.25357
Xu Y, Gao M, Zhang Y, Ning L, Zhao D, Ni Y (2022) Cellulose hollow annular nanoparticles prepared from high-intensity ultrasonic treatment. ACS Nano 16(6):8928–8938. https://doi.org/10.1021/acsnano.1c11167
Langan Nishiyama (2000) Wada, Sugiyama and Chanzy (2000) Crystal structure and hydrogen-bonding system in cellulose from neutron fiber diffraction. ABSTR PAP AMER CHEM SOC 219:U262–U262. https://doi.org/10.1021/ja0257319
Li L, Wu R, Guang S, Su X, Xu H (2013) The investigation of the hydrogen bond saturation effect during the dipole–dipole induced azobenzene supramolecular self-assembly. Phys Chem Chem Physics Pccp 15(47):20753–20763. https://doi.org/10.1039/c3cp52864b
Y. Li, YushangLiu, YangLai, WenchuanHuang, FengOu, AnpingQin, RuiLiu, XiangyangWang, Xu (2018) Ester crosslinking enhanced hydrophilic cellulose nanofibrils aerogel. ACS Sustain Chem Eng 6(9).https://doi.org/10.1021/acssuschemeng.8b02284
Funding
This work was financially supported by the Natural Science Basic Research Plan in Shaanxi Province of China (2021JQ-537) and Natural Science Foundation of Shaanxi University of Science & Technology.
Author information
Authors and Affiliations
Contributions
Tengfei Zhao: Writing—original draft, Software, Methodology, Data curation. Huaiqin Ma: Writing—review, Supervision, Project administration, Conceptualization. Lulu Ning: Writing—review & editing, Supervision, Project administration All authors contributed to the validation of the manuscript and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, T., Ma, H., Liu, Y. et al. Interfacial interactions between spider silk protein and cellulose studied by molecular dynamics simulation. J Mol Model 30, 156 (2024). https://doi.org/10.1007/s00894-024-05945-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05945-w