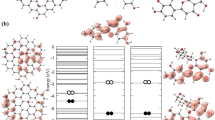
Abstract
Context
Heparin, one of the drugs reused in studies with antiviral activity, was chosen to investigate a possible blockade of the SARS-CoV-2 spike protein for viral entry through computational simulations and experimental analysis. Heparin was associated to graphene oxide to increase in the binding affinity in biological system. First, the electronic and chemical interaction between the molecules was analyzed through ab initio simulations. Later, we evaluate the biological compatibility of the nanosystems, in the target of the spike protein, through molecular docking. The results show that graphene oxide interacts with the heparin with an increase in the affinity energy with the spike protein, indicating a possible increment in the antiviral activity. Experimental analysis of synthesis and morphology of the nanostructures were carried out, indicating heparin absorption by graphene oxide, confirming the results of the first principle simulations. Experimental tests were conducted on the structure and surface of the nanomaterial, confirming the heparin aggregation on the synthesis with a size between the GO layers of 7.44 Å, indicating a C–O type bond, and exhibiting a hydrophilic surface characteristic (36.2°).
Methods
Computational simulations of the ab initio with SIESTA code, LDA approximations, and an energy shift of 0.05 eV. Molecular docking simulations were performed in the AutoDock Vina software integrated with the AMDock Tools Software using the AMBER force field. GO, GO@2.5Heparin, and GO@5Heparin were synthesized by Hummers and impregnation methods, respectively, and characterized by X-ray diffraction and surface contact angle.












Similar content being viewed by others
References
Hopkins J (2022) Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html
Wang D, Li Z, Liu Y (2020) An overview of the safety, clinical application and antiviral research of the COVID-19 therapeutics. J Infect Public Health 13:1405–1414. https://doi.org/10.1016/j.jiph.2020.07.004
Rhazouani A, Aziz K, Gamrani H et al (2021) Can the application of graphene oxide contribute to the fight against COVID-19? Antiviral activity, diagnosis and prevention. Curr Res Pharmacol Drug Discov 2:100062. https://doi.org/10.1016/j.crphar.2021.100062
Gupta I, Azizighannad S, Farinas ET, Mitra S (2021) Antiviral properties of select carbon nanostructures and their functionalized analogs. Mater Today Commun 29:102743. https://doi.org/10.1016/j.mtcomm.2021.102743
Seifi T, Reza Kamali A (2021) Antiviral performance of graphene-based materials with emphasis on COVID-19: a review. Med Drug Discov 11:100099. https://doi.org/10.1016/j.medidd.2021.100099
Shafiee A, Iravani S, Varma RS (2022) Graphene and graphene oxide with anticancer applications: challenges and future perspectives. MedComm (Beijing) 3. https://doi.org/10.1002/mco2.118
Dacrory S (2021) Antimicrobial Activity, DFT Calculations, and molecular docking of dialdehyde cellulose/graphene oxide film against Covid-19. J Polym Environ 29:2248–2260. https://doi.org/10.1007/s10924-020-02039-5
Fukuda M, Islam MS, Shimizu R et al (2021) Lethal interactions of SARS-CoV-2 with graphene oxide: implications for COVID-19 treatment. ACS Appl Nano Mater 4:11881–11887. https://doi.org/10.1021/acsanm.1c02446
Unal MA, Bayrakdar F, Nazir H et al (2021) Graphene oxide nanosheets interact and interfere with SARS-CoV-2 surface proteins and cell receptors to inhibit infectivity. Small 17:2101483. https://doi.org/10.1002/smll.202101483
Wang J, Yu Y, Leng T et al (2022) The inhibition of SARS-CoV-2 3CL Mpro by graphene and its derivatives from molecular dynamics simulations. ACS Appl Mater Interfaces 14:191–200. https://doi.org/10.1021/acsami.1c18104
Seabra AB, Paula AJ, de Lima R et al (2014) Nanotoxicity of graphene and graphene oxide. Chem Res Toxicol 27:159–168. https://doi.org/10.1021/tx400385x
Clausen TM, Sandoval DR, Spliid CB et al (2020) SARS-CoV-2 Infection depends on cellular heparan sulfate and ACE2. Cell 183:1043–1057.e15. https://doi.org/10.1016/j.cell.2020.09.033
Gupta Y, Maciorowski D, Zak SE et al (2021) Heparin: a simplistic repurposing to prevent SARS-CoV-2 transmission in light of its in-vitro nanomolar efficacy. Int J Biol Macromol 183:203–212. https://doi.org/10.1016/j.ijbiomac.2021.04.148
Mycroft-West CJ, Su D, Pagani I et al (2020) Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the Spike S1 receptor-binding domain with heparin. Thromb Haemost 120:1700–1715. https://doi.org/10.1055/s-0040-1721319
Tandon R, Sharp JS, Zhang F et al (2021) Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol 95. https://doi.org/10.1128/JVI.01987-20
Alzain AA, Elbadwi FA, Alsamani FO (2022) Discovery of novel TMPRSS2 inhibitors for COVID-19 using in silico fragment-based drug design, molecular docking, molecular dynamics, and quantum mechanics studies. Inform Med Unlocked 29:100870. https://doi.org/10.1016/j.imu.2022.100870
Ridgway H, Moore GJ, Mavromoustakos T et al (2022) Discovery of a new generation of angiotensin receptor blocking drugs: receptor mechanisms and in silico binding to enzymes relevant to SARS-CoV-2. Comput Struct Biotechnol J 20:2091–2111. https://doi.org/10.1016/j.csbj.2022.04.010
Ghosh K, SkA A, Gayen S, Jha T (2021) Chemical-informatics approach to COVID-19 drug discovery: exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J Mol Struct 1224:129026. https://doi.org/10.1016/j.molstruc.2020.129026
Ongtanasup T, Wanmasae S, Srisang S et al (2022) In silico investigation of ACE2 and the main protease of SARS-CoV-2 with phytochemicals from Myristica fragrans (Houtt.) for the discovery of a novel COVID-19 drug. Saudi. J Biol Sci 29:103389. https://doi.org/10.1016/j.sjbs.2022.103389
Gangadharan S, Ambrose JM, Rajajagadeesan A et al (2022) Repurposing of potential antiviral drugs against RNA-dependent RNA polymerase of SARS-CoV-2 by computational approach. J Infect Public Health 15:1180–1191. https://doi.org/10.1016/j.jiph.2022.09.007
Santos AF dos, Ortiz MM, Montagner GE, et al (2022) In-silico study of antivirals and non-antivirals for the treatment of SARS-COV-2. Disciplinarum Scientia 23:57–83. https://doi.org/10.37779/nt.v23i2.4200
Schultz JV, Tonel MZ, Martins MO, Fagan SB (2023) Graphene oxide and flavonoids as potential inhibitors of the spike protein of SARS-CoV-2 variants and interaction between ligands: a parallel study of molecular docking and DFT. Struct Chem. https://doi.org/10.1007/s11224-023-02135-x
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136:B864–B871. https://doi.org/10.1103/PhysRev.136.B864
Perdew JP, Zunger A (1981) Self-interaction correction to density-functional approximations for many-electron systems. Phys Rev B 23:5048–5079. https://doi.org/10.1103/PhysRevB.23.5048
ICMAB (2020) Instituto de Ciência de Materiais de Barcelona. https://departments.icmab.es/leem/siesta/
Soler JM, Artacho E, Gale JD et al (2002) The SIESTA method for ab initio order- N materials simulation. J Phys : Condensed Matter 14:2745–2779. https://doi.org/10.1088/0953-8984/14/11/302
García A, Papior N, Akhtar A et al (2020) Siesta : recent developments and applications. J Chem Phys 152:204108. https://doi.org/10.1063/5.0005077
Jauris IM, Matos CF, Saucier C et al (2016) Adsorption of sodium diclofenac on graphene: a combined experimental and theoretical study. Phys Chemist Chem Phys 18:1526–1536. https://doi.org/10.1039/C5CP05940B
Machado FM, Carmalin SA, Lima EC et al (2016) Adsorption of alizarin Red S dye by carbon nanotubes: an experimental and theoretical investigation. J Phys Chem C 120:18296–18306. https://doi.org/10.1021/acs.jpcc.6b03884
Jauris IM, Fagan SB, Adebayo MA, Machado FM (2016) Adsorption of acridine orange and methylene blue synthetic dyes and anthracene on single wall carbon nanotubes: a first principle approach. Comput Theor Chem 1076:42–50. https://doi.org/10.1016/j.comptc.2015.11.021
Hernández Rosas JJ, Ramírez Gutiérrez RE, Escobedo-Morales A, Chigo Anota E (2011) First principles calculations of the electronic and chemical properties of graphene, graphane, and graphene oxide. J Mol Model 17:1133–1139. https://doi.org/10.1007/s00894-010-0818-1
de Moraes EE, Tonel MZ, Fagan SB, Barbosa MC (2019) Density functional theory study of π-aromatic interaction of benzene, phenol, catechol, dopamine isolated dimers and adsorbed on graphene surface. J Mol Model 25:302. https://doi.org/10.1007/s00894-019-4185-2
Tournus F, Charlier J-C (2005) Ab initio study of benzene adsorption on carbon nanotubes. Phys Rev B 71:165421. https://doi.org/10.1103/PhysRevB.71.165421
Tournus F, Latil S, Heggie MI, Charlier J-C (2005) π-stacking interaction between carbon nanotubes and organic molecules. Phys Rev B 72:075431. https://doi.org/10.1103/PhysRevB.72.075431
Li B, Ou P, Wei Y et al (2018) Polycyclic aromatic hydrocarbons adsorption onto graphene: a DFT and AIMD study. Materials 11:726. https://doi.org/10.3390/ma11050726
Arrigoni M, Madsen GKH (2019) Comparing the performance of LDA and GGA functionals in predicting the lattice thermal conductivity of III-V semiconductor materials in the zincblende structure: the cases of AlAs and BAs. Comput Mater Sci 156:354–360. https://doi.org/10.1016/j.commatsci.2018.10.005
Cresti A, Lopez-Bezanilla A, Ordejón P, Roche S (2011) Oxygen surface functionalization of graphene nanoribbons for transport gap engineering. ACS Nano 5:9271–9277. https://doi.org/10.1021/nn203573y
Trott O, Olson AJ (2009) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem NA-NA. https://doi.org/10.1002/jcc.21334
Martins MO, Silva IZ da, Fagan SB, Santos AF dos (2021) Docking fundamentals for simulation in nanoscience. Disciplinarum Scientia 22:67–76. https://doi.org/10.37779/nt.v22i3.4106
Bell EW, Zhang Y (2019) DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J Cheminform 11:40. https://doi.org/10.1186/s13321-019-0362-7
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Dassault Systèmes (2023) Biovia Discovery Studio
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2020) AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol Direct 15:12. https://doi.org/10.1186/s13062-020-00267-2
van Dijk ADJ, Bonvin AMJJ (2006) Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics 22:2340–2347. https://doi.org/10.1093/bioinformatics/btl395
Pubchem (2022) Pubchem. https://pubchem.ncbi.nlm.nih.gov/compound/5288499. Accessed 2 Oct 2022
Chemcraft (2022) Chemcraft - graphical software for visualization of quantum chemistry computationsy. . https://www.chemcraftprog.com. Accessed 5 Dec 2022
PDB Protein Data Bank (2020) SARS-CoV-2 spike ectodomain structure (open state). https://www.rcsb.org/structure/6VYB
PDB Protein Data Bank (2022) SARS-CoV-2 Omicron BA.4 variant spike. https://www.rcsb.org/structure/7XNQ
Hummers WS, Offeman RE (1958) Preparation of graphitic oxide. J Am Chem Soc 80:1339–1339. https://doi.org/10.1021/ja01539a017
Moraes FC, Rossi B, Donatoni MC et al (2015) Sensitive determination of 17β-estradiol in river water using a graphene based electrochemical sensor. Anal Chim Acta 881:37–43. https://doi.org/10.1016/j.aca.2015.04.043
Ahmad W, Mehmood U, Al-Ahmed A et al (2016) Synthesis of zinc oxide/titanium dioxide (ZnO/TiO2) nanocomposites by wet incipient wetness impregnation method and preparation of ZnO/TiO2 paste using poly(vinylpyrrolidone) for efficient dye-sensitized solar cells. Electrochim Acta 222:473–480. https://doi.org/10.1016/j.electacta.2016.10.200
Le GTT, Chanlek N, Manyam J et al (2019) Insight into the ultrasonication of graphene oxide with strong changes in its properties and performance for adsorption applications. Chem Eng J 373:1212–1222. https://doi.org/10.1016/j.cej.2019.05.108
Silva CC, Graça MPF, Valente MA, Sombra ASB (2007) Crystallite size study of nanocrystalline hydroxyapatite and ceramic system with titanium oxide obtained by dry ball milling. J Mater Sci 42:3851–3855. https://doi.org/10.1007/s10853-006-0474-0
De Rosa C, Auriemma F (2013) Crystals and crystallinity in polymers: diffraction analysis of ordered and disordered crystals. Wiley
Bursulaya BD, Totrov M, Abagyan R, Brooks III CL (2003) Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des 17:755–763. https://doi.org/10.1023/B:JCAM.0000017496.76572.6f
Krishnamoorthy K, Veerapandian M, Yun K, Kim S-J (2013) The chemical and structural analysis of graphene oxide with different degrees of oxidation. Carbon N Y 53:38–49. https://doi.org/10.1016/j.carbon.2012.10.013
Siburian R, Sihotang H, Lumban Raja S et al (2018) New route to synthesize of graphene nano sheets. Oriental Journal of Chemistry 34:182–187. https://doi.org/10.13005/ojc/340120
Bureau MN, Denault J, Cole KC, Enright GD (2002) The role of crystallinity and reinforcement in the mechanical behavior of polyamide-6/clay nanocomposites. Polym Eng Sci 42:1897–1906. https://doi.org/10.1002/pen.11082
Kim B-H, Lee S-H, Han S-W et al (2016) Anatomy of high recyclability of graphene oxide based palladium nanocomposites in the Sonogashira reaction: on the nature of the catalyst deactivation. Mater Express 6:61–68. https://doi.org/10.1166/mex.2016.1280
Kang W-S, Rhee KY, Park S-J (2017) Influence of surface energetics of graphene oxide on fracture toughness of epoxy nanocomposites. Compos B Eng 114:175–183. https://doi.org/10.1016/j.compositesb.2017.01.032
Fu J, Ji J, Yuan W, Shen J (2005) Construction of anti-adhesive and antibacterial multilayer films via layer-by-layer assembly of heparin and chitosan. Biomaterials 26:6684–6692. https://doi.org/10.1016/j.biomaterials.2005.04.034
Pietsch T, Gindy N, Fahmi A (2009) Nano- and micro-sized honeycomb patterns through hierarchical self-assembly of metal-loaded diblock copolymer vesicles. Soft Matter 5:2188–2197. https://doi.org/10.1039/B814061H
Zheng Y, Zaoui A (2017) Wetting and nanodroplet contact angle of the clay 2:1 surface: the case of Na-montmorillonite (001). Appl Surf Sci 396:717–722. https://doi.org/10.1016/j.apsusc.2016.11.015
Tang K, Yu J, Zhao Y et al (2006) Fabrication of super-hydrophobic and super-oleophilic boehmite membranes from anodic alumina oxide film via a two-phase thermal approach. J Mater Chem 16:1741. https://doi.org/10.1039/b600733c
Acknowledgements
Acknowledgment for computational support from CENAPAD-SP (National Center for High-Performance Processing in São Paulo).
Funding
This work was carried out with the support of the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brazil (CAPES) Financing Code 001, TELEMEDICINA 1690389P, INCT Nanomateriais de Carbono (CNPq).
Author information
Authors and Affiliations
Contributions
AFS, MOM, DMD, JSL, CBD, and MZT developed the conception and design of the study. AKM, CBD, and DVL developed the methodology and are conducting safety and efficacy tests with cells. DMD, SRM, and MSF developed the methodology and performed compound characterization tests. AFS, MOM, MZT, JSL, and JOA performed computer simulations. AFS, MZT, JOA, CBD, AKM, and DVS reviewed the images and wrote the article under the supervision of IZS, SF, and SRM. AFS, JOA, JSL, and DMD reviewed the results. All authors reviewed and commented on the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This paper belongs to the Topical Collection IX Symposium on Electronic Structure and Molecular Dynamics – IX SeedMol
Supplementary Material
Video 1 of version 01 of the ab initio simulation results. (MPG 1339 kb)
Video 2 of version 02 of the ab initio simulation results. (MPG 3968 kb)
Video 3 of version 03 of the ab initio simulation results. (MPG 870 kb)
Video 4 of version 04 of the ab initio simulation results. (MPG 4700 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
dos Santos, A.F., Martins, M.O., Lameira, J. et al. Evaluation interaction of graphene oxide with heparin for antiviral blockade: a study of ab initio simulations, molecular docking, and experimental analysis. J Mol Model 29, 235 (2023). https://doi.org/10.1007/s00894-023-05645-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05645-x