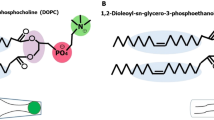
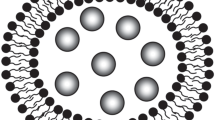
Abstract
In this study, liposome and transfersome were successfully constructed using molecular dynamics simulation. Three drugs with different polarity, including 5-fluorouracil, ligustrazine, and osthole, were selected as model drugs to study the distribution of drugs in lipid vesicles by calculating the radial distribution function and the potential of mean force. The solubility parameters between drugs and different regions in lipid vesicles were calculated to characterize the compatibility of drugs in different regions in lipid vesicles, which provided the basis for the conclusion of this paper. It showed that the radial distribution function and the potential of mean force were consistent in the characterization of drug distribution in vesicles, and the drug distribution in vesicles was closely related to the compatibility between drugs and vesicles. Therefore, the radial distribution function and the potential of mean force can be used to characterize the distribution of drugs in vesicles, and molecular simulation technology has a great potential in studying the characteristics of vesicles.

Graphical abstract











Similar content being viewed by others
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Fahr A, Van Hoogevest P, Kuntsche J, Leigh ML (2016) Lipophilic drug transfer between liposomal and biological membranes: what does it mean for parenteral and oral drug delivery? J Liposome Res 16(3):281–301. https://doi.org/10.1080/08982100600848702
Shishir MRI, Karim N, Gowd V, Zheng XD, Chen W (2019) Liposomal delivery of natural product: a promising approach in health research. Trends Food Sci Technol 85:177–200. https://doi.org/10.1016/j.tifs.2019.01.013
Sheikhpour M, Barani L, Kasaeian A (2017) Biomimetics in drug delivery systems: a critical review. J Control Release 253:97. https://doi.org/10.1016/j.jconrel.2017.03.026
Suri R, Neupane YRR, Kohli K, Gaurav KJ (2020) Polyoliposomes: novel polyol modified lipidic nanovesicles for dermal and transdermal delivery of drugs. Nanotechnology 31(35). https://doi.org/10.1088/1361-6528/ab912d
Natsheh H, Touitou E (2020) Phospholipid vesicles for dermal/transdermal and nasal administration of active molecules: the effect of surfactants and alcohols on the fluidity of their lipid bilayers and penetration enhancement properties. Molecules. https://doi.org/10.3390/molecules25132959
Huang H, Yang XR, Li HL, Lu HS, Oswald J, Liu YM, Zeng J, Jin CH, Peng XC, Liu JY (2020) iRGD decorated liposomes: a novel actively penetrating topical ocular drug delivery strategy. Nano Res. https://doi.org/10.1007/s12274-020-2980-9
Yoshikawa N, Fumoto S, Nakashima M, Shimokawa K, Miyamoto H, Nishida K (2013) The role of Fibronectin in pulmonary gene transfer following intravenous administration of lipoplex in mice. Biol Pharm Bull 36(11):1807–1813. https://doi.org/10.1007/s12274-020-2980-9
Czajkowska-Kosnik A, Szekalska M, Winnicka K (2019) Nanostructured lipid carriers: a potential use for skin drug delivery systems. Pharmacol Rep 71(1):156–166. https://doi.org/10.1016/j.pharep.2018.10.008
Opatha SAT, Titapiwatanakun V, Chutoprapat R (2020) Transfersomes: a promising nanoencapsulation technique for transdermal drug delivery. Pharmacol Pharm. https://doi.org/10.3390/pharmaceutics12090855
Wu Z, Yang C, Chen L, Chen L, Ma L (2019) A multiscale study on the effect of sodium cholate on the deformation ability of elastic liposomes. AAPS PharmTech 20(8). https://doi.org/10.1208/s12249-019-1485-x
Verma S, Utreja P (2019) Vesicular nanocarrier based treatment of skin fungal infections: potential and emerging trends in nanoscale pharmacotherapy. Asian J Pharm Sci 14(2):117–129. https://doi.org/10.1016/j.ajps.2018.05.007
Mehta SK, Jindal N (2013) Mixed micelles of lecithin-tyloxapol as pharmaceutical nanocarriers for anti-tubercular drug delivery. Colloid Surf B 110:419–425. https://doi.org/10.1016/j.colsurfb.2013.05.015
Barone A, Cristiano MC, Cilurzo F, Locatelli M, Iannotta D, Di Marzio L, Paolino D (2020) Ammonium glycyrrhizate skin delivery from ultradeformable liposomes: a novel use as an anti-inflammatory agent in topical drug delivery. Colloid Surf B 193. https://doi.org/10.1016/j.colsurfb.2020.111152
Imkan Ali I, Ullah S, Imran M, Saifullah S, Hussain K, Shah MR (2020) Synthesis of biocompatible triazole based non-ionic surfactant and its vesicular drug delivery investigation. Chem Phys Lipids 228:9. https://doi.org/10.1016/j.chemphyslip.2020.104894
Bombelli C, Caracciolo G, Di Profio P, Diociaiuti M, Luciani P, Mancini G, Venanzi M (2005) Inclusion of a photosensitizer in liposomes formed by DMPC/Gemini surfactant: correlation between physicochemical and biological features of the complexes. J Med Chem 48(15):4882–4891. https://doi.org/10.1021/jm050182d
Aloisio C, Antimisiaris SG, Longhi MR (2017) Liposomes containing cyclodextrins or meglumine to solubilize and improve the bioavailability of poorly soluble drugs. J Mol Liq 229:106–113. https://doi.org/10.1016/j.molliq.2016.12.035
Truszkowski A, Epple M, Fiethen A, Zielesny A, Kuhn H (2013) Molecular fragment dynamics study on the water-air interface behavior of non-ionic polyoxyethylene alkyl ether surfactants. J Colloid Interface Sci 410:140–145. https://doi.org/10.1016/j.jcis.2013.07.069
Park KL (2017) Emergence of hydrogen bonds from molecular dynamics simulation of substituted N-phenylthiourea-catechol oxidase complex. Arch Pharm Res 40(1):57–68. https://doi.org/10.1007/s12272-016-0866-x
Higashi H, Oda T, Iwai Y, Arai Y (2004) Calculation of diffusion coefficients for carbon dioxide plus solute system near the critical conditions by non-equilibrium molecular dynamics simulation. Fluid Phase Equilib 219(1):55–60. https://doi.org/10.1016/j.fluid.2004.01.014
General IJ, Meirovitch H (2013) Absolute free energy of binding and entropy of the FKBP12-FK506 complex: effects of the force field. J Chem Theory Comput 9(10):4609–4619. https://doi.org/10.1021/ct400484u
Hashemzadeh H, Javadi H, Darvishi MH (2020) Study of structural stability and formation mechanisms in DSPC and DPSM liposomes: a coarse-grained molecular dynamics simulation. Sci Rep. https://doi.org/10.1038/s41598-020-58730-z
Carpenter TS, López CA, Neale C, Montour C, Ingolfsson HI, Di Natale F, Lightstone FC, Gnanakaran S (2018) Capturing phase behavior of ternary lipid mixtures with a refined martini coarse-grained force field. J Chem Theory Comput. https://doi.org/10.1021/acs.jctc.8b00496
Chang Y, Xinging D, Shufang Y, Lina M, Liping C, Ruilin G, Xiaowen W (2019) Coarse-grained molecular dynamics simulations of the effect of edge activators on the skin permeation behavior of transfersomes. Colloid Surf B. https://doi.org/10.1016/j.colsurfb.2019.110462
Santiago R, Reigada R (2019) Interaction modes between nanosized graphene flakes and liposomes: adsorption, insertion and membrane fusion. Biochim Biophys Acta Gen Subj 1863(4):723–731. https://doi.org/10.1016/j.colsurfb.2019.110462
Lyubartsev AP (2005) Multiscale modeling of lipids and lipid bilayers. Eur Biophys J Biophys 35(1):53–61. https://doi.org/10.1007/s00249-005-0005-y
Leekumjorn S, Sum AK (2007) Molecular characterization of gel and liquid-crystalline structures of fully hydrated POPC and POPE bilayers. J Phys Chem B 111(21):6026–6033. https://doi.org/10.1021/jp0686339
Suits F, Pitman MC, Feller SE (2005) Molecular dynamics investigation of the structural properties of phosphatidylethanolamine lipid bilayers. J Chem Phys 122(24):9. https://doi.org/10.1063/1.1899152
Kasson PM, Pande VS (2007) Control of vesicle fusion by changes in membrane properties: predictions from molecular dynamics simulation. Biophys J:182A–182A
Nakagawa KM, Noguchi H (2015) Morphological changes of amphiphilic molecular assemblies induced by chemical reactions. Soft Matter 11(7):1403–1411. https://doi.org/10.1039/c4sm02571g
Lundborg M, Narangifard A, Wennberg CL, Lindahl E, Daneholt B, Norlen L (2018) Human skin barrier structure and function analyzed by cryo-EM and molecular dynamics simulation. J Struct Biol 203(2):149–161. https://doi.org/10.1016/j.jsb.2018.04.005
Jalili S, Saeedi M (2016) Study of curcumin behavior in two different lipid bilayer models of liposomal curcumin using molecular dynamics simulation. J Biomol Struct Dyn 34(2):327–340. https://doi.org/10.1080/07391102.2015.1030692
Wood I, Fabian L, Moglioni A, Cabeca LF, de Paula E, Pickholz M (2019) Combining nuclear magnetic resonance with molecular dynamics simulations to address sumatriptan interaction with model membranes. Chem Phys Lipids 225:9. https://doi.org/10.1016/j.chemphyslip.2019.104792
El-Alim SHA, Kassem AA, Basha M, Salama A (2019) Comparative study of liposomes, ethosomes and transfersomes as carriers for enhancing the transdermal delivery of diflunisal: in vitro and in vivo evaluation. Int J Pharm. https://doi.org/10.1016/j.ijpharm.2019.04.001
Arnarez C, Uusitalo JJ, Masman MF, Ingolfsson HI, de Jong DH, Melo MN, Periole X, de Vries AH, Marrink SJ (2015) Dry Martini, a coarse-grained force field for lipid membrane simulations with implicit solvent. J Chem Theory Comput. https://doi.org/10.1021/ct500477k
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2010) GROMACS: fast, flexible, and free. J Chem Theory Comput 6:3713–3720. https://doi.org/10.1021/ct100494z
Lemaalem M, Ahfir R, Derouiche A (2020) Static and dynamic properties of decane/water microemulsions stabilized by cetylpyridinium chloride cationic surfactant and octanol cosurfactant. RSC Adv. https://doi.org/10.1039/D0RA06313D
Warren DB, King D, Benameur H, Pouton CW, Chalmers DK (2013) Glyceride lipid formulations: molecular dynamics modeling of phase behavior during dispersion and molecular interactions between drugs and excipients. Pharm Res-Dordr 30(12):3238–3253. https://doi.org/10.1007/s11095-013-1206-1
Seifollah J, Marzieh S (2015) Study of curcumin behavior in two different lipid bilayer models of liposomal curcumin using molecular dynamics simulation. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2015.1030692
Acknowledgements
First of all, I would like to express my sincere gratitude to all those who have lent me hands in the course of writing this paper. Secondly, I would like to take this opportunity to show my thanks to my supervisor, Ms. Shi, who has given me so much useful advice on my writing and has tried her best to improve my paper. And I would like to express my gratitude to Ms. Dai who offered me references and information on time. Last but not the least, I would like to thank those leaders, teachers, and working staff at Beijing University of Chinese Medicine. Without their help, it would be much harder for me to finish my study and this paper.
Code availability
The experiment was carried out on the basis of Gromacs 4.6.3, Packmol 18.013, Visual Molecular Dynamics 1.9.2, and Materials Studio 7.0.
Funding
This work was funded by the Young Teachers Program of Beijing University of Chinese Medicine (Grant 2019-JYB-JS-016) and the Natural Science Foundation of Beijing, China (Grant No.7162122). All simulations were performed at the National Supercomputer Center in Guangzhou.
Author information
Authors and Affiliations
Contributions
We declare that this work was done by the authors named in this article, and all liabilities pertaining to claims relating to the content of this article will be borne by the authors. In addition, a declaration of the role of each author mentioned as follows: Dr. Xinyuan Shi and Mrs. Xingxing Dai contributed to the conception of the study; Mrs. Xiaowen Wu performed the experiment and acquired the data; Mrs. Yuyao Liao participated in the analysis of the results; Mrs. Mengke Sheng contributed significantly to the validation of data and visualization of results; Mrs. Xiaowen Wu aided in drafting and revising the manuscript; Dr. Xinyuan Shi and Mrs. Xingxing Dai helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
for publication.
Corresponding author
Ethics declarations
Consent to participate
Using two commonly used vesicles and three representative drugs as models, the simulated force field was fully validated.
Consent for publication
This study provides a new evaluation method for the drug loading location in the vesicle and explains the related mechanism.
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 238 kb)
Rights and permissions
About this article
Cite this article
Wu, X., Dai, X., Liao, Y. et al. Investigation on drug entrapment location in liposomes and transfersomes based on molecular dynamics simulation. J Mol Model 27, 111 (2021). https://doi.org/10.1007/s00894-021-04722-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04722-3