Abstract
The physical properties of lipid bilayers are sensitive to the specific type and composition of the lipids that make up the many different types of cell membranes. Studying model bilayers of representative heterogeneous compositions can provide key insights into membrane functionality. In this work, we use atomistic molecular dynamics simulations to characterize key properties in a number of bilayer membranes of varying composition. We first examine basic properties, such as lipid area, volume, and bilayer thickness, of simple, homogeneous bilayers comprising several lipid types, which are prevalent in biological membranes. Such lipids are then used in simulations of heterogeneous systems representative of bacterial, mammalian, and cancer membranes. Our analysis is especially focused on depth-dependent, transmembrane profiles; in particular, we calculate lateral pressure and dipole potential profiles, two fundamental properties which play key roles in a large number of biological functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipid bilayers are fundamental structures in biology; they form a continuous barrier around cells, compartmentalize the intracellular space, act as selective barriers for the permeation of molecules, and also play key roles in modulating the dynamics, organization, and function of membrane proteins [1,2,3,4]. A detailed understanding of the properties of lipid bilayers is central to biology, and can help the advancement of related research areas such as biomedicine and drug design [5,6,7]. Molecular dynamics (MD) simulations provide a powerful, complementary approach to experiments, and as such they have been used to study extensively many aspects of lipid bilayers [8,9,10,11,12,13,14]. Simulations are especially useful to study those properties that fluctuate substantially as a function of depth inside the bilayer, as experimental investigation is very arduous, mostly because of the small thickness (∼45 Å), high fluidity and heterogeneity of typical bilayers.
In this study, we use atomistic MD simulations to characterize a number of fundamental physical properties for various bilayers of different lipid compositions. In particular, we investigate multicomponent, asymmetric lipid bilayers with complex compositions mimicking mammalian, cancer, and bacterial plasma membranes.
The mammalian plasma membranes are typically made of approximately 65% glycerolipids, 10% sphingolipids, and 25% sterols, and have a highly asymmetric lipid composition [15,16,17]. Specifically, the extracellular leaflet is mostly composed of phosphatidylcholine (PC) lipids such as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and of sphingolipids such as sphingomyelin and glycosphingolipids. The intracellular leaflet on the other hand is enriched in phosphatidylethanolamine (PE) lipids such as 1- palmatoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine(POPE) and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPE), and in negatively charged phosphatidylserine (PS) lipids such as 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS) and 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS). While many phospholipids may show no preference to a specific leaflet of the membrane, PS lipids are known to be predominantly located in the inner leaflets of the cells [18,19,20,21]. A consequence of this irregular lipid composition is that the inner leaflet of the mammalian plasma membrane is anionic in nature [15, 16].
When healthy cells develop into cancer ones, the transformation is accompanied by a 5–9 times increase in PS lipids in the external leaflet; in fact, this change has been the primary mode of recognition of transformed cells by monocytes as part of the body’s immune response [22, 23]. Indeed, in many studies, efforts have been made to exploit this expression of PS lipids in external leaflets to design cationic antimicrobial peptides (AMPs) and anticancer peptides (ACPs) that specifically target tumor cells, without harming the surrounding normal cells [24, 25]. Another alteration found in tumor cells is the reduction of cholesterol. Cholesterol is a major component of all eukaryotic membranes, where its presence induces increased ordering of lipid hydrocarbon chains [26,27,28], decreased lateral diffusion of lipids along the membrane plane [29] and increased lateral pressure profile magnitudes [30, 31].
In this study, we simulate both a mammalian and a cancer bilayer membrane models based on the representative compositions of Klahn et al. [32], which reflect the two aforementioned features of healthy mammalian cells and transformed cancer ones. It is important to stress that the models used in our work include only representative lipids of a limited number of prevalent species, and thus do not capture the full diversity of lipids in real cells; for example, predominant species such as polyunsaturated lipids, as well as PA, PI/PIPs and glycolipids, are not included. Especially considering the heterogeneity in saturation levels of lipid tails in real cells, the presence of lipids with only monounsaturated tails is a limitation of our study.
Regarding the bacterial membrane, we have approximated the complexity of real systems [33] with a model consisting of a 2:1 POPE:POPG lipid composition, which is arguably representative of the fundamental character of most Gram-negative bacterial membranes. In fact, the lipid composition used in our work has been used previously both in experiments [34] and MD simulations [35, 36]. In particular, the PG lipids induce a net negative charge on bacterial membranes (similarly to the effect of PS lipids on cancer membranes).
Apart from the three complex bilayers discussed above (mammalian, cancer, and bacterial), we also simulated homogeneous bilayers composed of individual lipids present in the complex systems, in order to investigate their contributions to the various properties analyzed. For each bilayer, we first calculate a number of basic membrane structural properties, such as area and volume per lipid, and bilayer thickness. We then place special focus on the transmembrane profiles of lateral pressure and dipole potential, as these fundamental properties, which vary significantly as a function of depth inside the membrane, are believed to play important roles in many membrane processes [37,38,39].
Considering the types of known interactions that exist at various depths inside the lipid membrane, an internal inhomogeneous distribution of lateral pressure is expected theoretically and can be predicted qualitatively [40]. Experiments have also been conducted to quantify the lateral pressure using pyrene moieties as probes to detect the local pressure change [41, 42]. However, only relative pressure changes in the tail regions could be obtained, and the accuracy of the experimental data is disputed [12, 41]. More recently, numerous molecular simulations have provided quantitative evidence that the lateral pressure profile is characterized by depth-dependent pressure variations of the order of hundreds of atmospheres [12, 40, 43]. Furthermore, it is believed that the corresponding forces affect embedded proteins and permeating molecules, with repercussions on many membrane phenomena [1, 39, 44, 45]. For example, specific changes in the membrane lateral pressure may underpin the basic mechanism of general anesthesia by controlling the opening of ion channels that conduct neural signals [46]. An important factor that determines the shape and magnitude of the lateral pressure profile is the specific composition of the lipid bilayer. Notably, it has been shown that even small changes in the lipid composition can induce large variations in the pressure profiles [39]. Previous simulation studies have focused on the effects of cholesterol [30, 31], changing unsaturation levels of constituent lipids [47, 48] as well as the lamellar vs. nonlamellar lipid ratio using coarse-grained [49] and atomistic models [50]. Perrin et al. [51] have reported the pressure profiles for a 1:1 POPE:POPG, which is similar to our 2:1 POPE:POPG bacterial bilayer. However, as far as we are aware, no previous MD study has been reported on pressure profiles for the complex mammalian and cancer bilayer systems considered in this work.
The dipole potential is an intramembrane electrostatic potential which arises at the water–lipid interface due to the ordering of water molecular dipoles as well as dipolar charge distributions on lipid molecules [37, 52]. A direct measurement of the dipole potential would require the deployment of nanoscale electrodes at different depths within the bilayer, which has proved so far practically impossible [52]. It has been, however, possible to estimate the potential difference between the bilayer center and the outer water phase through indirect experimental measurements. In a pioneering study on ion transport [53], the transbilayer conductivity of two fat-soluble ions with similar sizes was observed to be drastically different, with the permeability of the negative ion (TPB-) being about 105 times higher than the positive ion (TPP+). This difference was ascribed to the existence of a positive electrostatic potential within the membrane. Some follow-up studies using varied methods and experiments confirmed the presence of such an electrostatic potential and estimated a magnitude of several hundreds of mV [52, 54]. It is crucial to note that the dipole potential fluctuates sharply within short distances of 2–3 nm, which is the approximate thickness of each of the monolayers in a typical bilayer. The corresponding electric field (which corresponds to the spatial derivative of the dipole potential) can therefore reach extremely large values, predicted to be in the range of 107-109 V/m. As a result, many electrostatically driven phenomena, such as binding and permeation of charged or polar molecules, as well as dynamics and interactions of ligands and corresponding transmembrane proteins, may be affected by changes in dipole potential. Past studies have considered the effects of lipid composition on the dipole potential, with a focus on the headgroup type [55], ether or ester linkage type [56] and double bonds in the chain region [57]. An atomistic MD simulation study by Ding et al. [50] focused on the effects of altering the lamellar vs. nonlamellar lipid composition. Overall, previous studies have considered relatively simple systems, either homogeneous bilayers or comprising two lipid types. In this work, we report for the first time the dipole potential of more complex and biologically relevant bacterial, mammalian, and cancer model membranes.
Methods
Lipid bilayer systems
Details of the composition of the lipid bilayer systems simulated in this work are reported in Table 1.
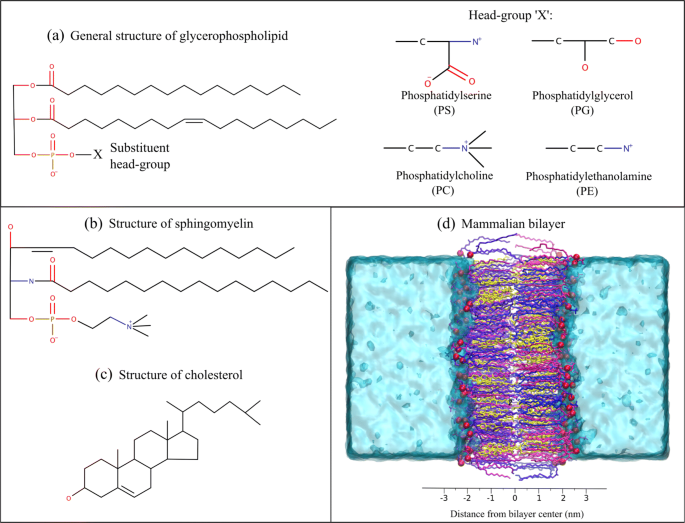
In particular, we study four homogeneous systems, each containing a single lipid type: POPC, POPE, POPG, and POPS. All the PC, PE, PG and PS lipids in this work have 1-phosphatidyl-2-oleoyl (PO) tails. The POPC system is then extended to include extra components, resulting in three mixed systems of incremental complexity, which include cholesterol (Chol) and palmitoylsphingomyelin (PSM): PC:Chol, PC:Chol:PSM and PC:Chol:PSM:PE. Palmitoylsphingomyelin (PSM) consists of 18:1-sphingosine and 16:0-palmitic acid. The molecular structures of all the lipid types simulated in this study are displayed in Fig. 1. Three additional systems are considered as models for bacterial, mammalian, and cancer membranes. Specifically, the bacterial membrane consists of a 2:1 PE:PG composition, following Horn et al. [58]. Regarding the mammalian and cancer membranes, their composition was taken from Klahn et al. [32]; supplementary Fig. S1 displays the percentage lipid composition and Table 2 reports their asymmetric lipid distribution in the outer and inner leaflets.
Molecular structures of all a glycerophospholipids, b palmitoylsphingomyelin and c cholesterol used in this study. Explicit hydrogens are omitted for clarity. d A representative snapshot of the mammalian bilayer system (created in VMD [108]) simulated in this study, with an approximate scale of distance from the bilayer center. Different colors correspond to different lipid species. The lipid tails are displayed in licorice, phosphate atoms with VDW, and water molecules with QuickSurf representation
All systems were fully hydrated with water and were neutralized with counter-ions wherever necessary. The CHARMM36 force field and the corresponding TIP3P implementation were used to model the lipids and water, respectively [59, 60]. System configurations were set up using Membrane Builder [61,62,63] from the Charmm-GUI [64]. In general, when constructing asymmetric bilayers, it is important to include the correct number of lipids in each leaflet, as any mismatch could introduce unrealistic effects, such as non-zero leaflet tension [65]. As a check on our systems, we carried out a number of extra simulations of symmetric bilayers, each comprising a lipid composition taken from each of the leaflets in the asymmetric systems. We then calculated the XY leaflet areas and compared them. As shown in the Supplementary Information (Fig. S3), after an initial equilibration the symmetric systems converge to areas that are at most ∼ 3% different than the corresponding asymmetric systems. Such a difference, which is expected to correspond to 3–4 lipids at most, is arguably negligible and unlikely to significantly affect the bilayer properties considered here.
The simulation input files are publicly available on figshare.com, see the Supplementary Information for the specific URL of each system.
Simulation details
Molecular dynamics simulations were performed using Gromacs version 5.1 [66, 67]. All the controls and bacterial systems were subjected to energy minimization and then NPT equilibration for 1 ns. The systems were coupled to the velocity rescale thermostat [68] with a coupling time constant of 0.1 ps. Except for the POPE, all the other systems were simulated at 303 K. The POPE bilayer was simulated at 310 K for consistency with the updated CHARMM36 force field [60]; such a slightly higher temperature typically used in previous simulations of the CHARMM36 model for POPE [60, 69] reproduces the correct liquid phase experimental behavior while preventing the formation of a gel phase (this is depicted in Fig. ?? in the Supplementary Information). Semi-isotropic pressure coupling [70,71,72] was carried out using the Berendsen barostat [73] for equilibration and the Parrinello–Rahman barostat [74] for production runs, with a coupling time constant of 2 ps. The SETTLE algorithm [75] was used to constrain bonds and angles in water molecules. All other hydrogen-related bonds were constrained using the LINCS algorithm [76], with two iterations in every step for correcting rotational effects and a numerical expansion up to the fourth order. Volume compressibility was chosen to be 4.5 ⋅ 10− 5 bar-1. For short-range van der Waals and electrostatic cutoff, the default optimized distance of 1.2 nm was used and a force-switch potential modifier was applied from 1 to 1.2 nm. Long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) algorithm [77]. The neighbor list was updated using grid-based search procedure every five steps. Each system was simulated for 1 μs; means and standard errors for the properties of interest were calculated using three trajectory blocks, corresponding to the intervals of 251–500, 501–750, and 751–1000 ns, while the initial 250 ns was regarded as equilibration.
Some extra steps were taken for equilibrating the mammalian, cancer, and the intermediate systems (PC:Chol:PSM and PC:Chol:PSM:PE) due to their complexity. In particular, these systems were subjected to stepwise energy minimization and equilibration under the NVT and NPT ensembles. The systems were coupled to the Berendsen thermostat at 303 K with a coupling time constant of 1 ps. The pressure was maintained at 1 atm using semi-isotropic pressure coupling with the Berendsen barostat [73]. For production runs, the thermostat and barostat were switched to velocity rescale and Parrinello–Rahman respectively, consistently with the other systems.
Calculations and data analysis
Structural properties
Following a common convention, the lipid bilayers were set up along the XY -plane, with the Z-axis thus crossing the membrane perpendicularly. The area per lipid (AL) is defined as the cross-sectional area (AXY) of the whole system along the bilayer surface plane (XY -plane), divided by half the total number of lipids (NL) present in the bilayer, i.e., AL = AXY/(NL/2). An exception to this method, however, is the PC:Chol bilayer, for which the area per lipid for POPC in the presence of cholesterol was calculated using the freely available tool APL@VORO [78]. The volume per lipid (VL) was calculated from the total volume of the simulation box (Vbox) as VL = (Vbox − Vwater)/NL, where Vwater is calculated from simulation of a pure water box at 303 K (at 310 K for POPE bilayer) and 1 atm. The bilayer thickness (dHH) was calculated as the head-to-head distance between the two peaks of the electron density profile. The electron density profiles were calculated using the gmx density tool in Gromacs, with the simulation box divided into 500 slabs parallel to the XY -plane.
Lateral pressure profile
To calculate the lateral pressure profile π(z) we used GROMACS-LS [79,80,81], a custom version of GROMACS that calculates the local stress tensor in 3D. This package was used to rerun the simulated trajectories and output the local stress tensors. Since the long-range electrostatic solver is not implemented in GROMACS-LS, an increased cutoff distance of 2.0 nm was used for Coulomb interactions, as recommended by the package developers. The simulation box was divided into 0.1 Å thick slabs parallel to the XY -plane, with the lateral pressure π(zs) at slab s defined as:
where Pxx(zs), Pyy(zs) and Pzz(zs) are the diagonal elements of the pressure tensor for slab s, along the X, Y and Z axes respectively. All the profiles have been smoothed by adaptive high-order spline fitting for clarity [82].
Dipole potential
We calculated the transmembrane dipole potential ψ(z) using the Gromacs tool gmx potential. In particular, the potential is obtained by summing the charge density distribution ρ(z) per slab, where the simulation box was divided into 500 slabs of ∼0.18 Å thickness each. The dipole potential is obtained by double integration using the following expression [71]:
where 𝜖0 is the electrostatic permittivity in vacuum.
Results and discussion
Structural properties
Simulation results for area per lipid AL, volume per lipid VL, and bilayer thickness dHH are reported in Table 3, together with available literature data from experiments and previous atomistic simulations.
For the homogeneous bilayers, as well as the PC:Chol system, our results are in good agreement with the values previously reported in the literature, thus validating our simulation setup.
The bacterial bilayer, being composed of POPE and POPG lipids in the ratio 2:1, exhibits intermediate values for its structural properties, although closer to the homogeneous POPE values. This is expected, due to POPE’s higher concentration in the bacterial bilayer, however it appears that POPE’s contribution is amplified in relation to its concentration. In particular, the bacterial bilayer exhibits condensation, in that the lipid area for the two-lipid mixture is lower than expected by simply taking a weighted average over the individual lipid areas of the POPE and POPG bilayers. This effect is also reflected in the electron density profiles, reported in Fig. 2. Specifically, it can be seen that the bacterial profile exhibits peaks that are larger than the constituent lipid profiles.
The mammalian lipid bilayer, which is a complex mixture of five different types of lipids, exhibits the lowest area and volume per lipid (∼42 Å2 and ∼989 Å3, respectively) and the highest bilayer thickness of ∼47 Å. This can be attributed primarily to the condensation effect induced by the predominance of cholesterol (34%) in the bilayer. A number of studies have indeed demonstrated that the presence of cholesterol induces a reduction in AL and VL and an increase in dHH in phospholipid bilayers [83,84,85,86,87]. Our own PC:Chol bilayer (AL: ∼45 Å2), with one-third cholesterol content shows a ∼30% reduction in AL as compared to POPC (∼63 Å2), in close agreement with such previous studies [83,84,85,86,87]. Compared to the mammalian bilayer, the cancer composition is characterized by a decrease in cholesterol and an increase in PS and PE lipids (also see supplementary Fig. S1). Such changes bring about an ∼18% increase in AL, from ∼42 Å2 to ∼46 Å2, an ∼10% increase in VL, from ∼990 Å3 to ∼1085 Å3, and a ∼3% decrease in bilayer thickness, from ∼47.3 Åto ∼45.9 Å. The presence of cholesterol in lipid bilayers is known to increase the ordering of lipid chains by intercalating in free spaces between them [88], effectively decreasing the lipid area; since a lipid membrane behaves as an incompressible fluid [86], this decrease in the area per lipid causes the bilayer thickness to increase. The difference in lipid composition between mammalian and cancer bilayers is also reflected in the electron densities, as illustrated in Fig. 3. It can be noted, in particular, that the mammalian profile shows a broadening of its peaks in the headgroup region compared to its cancer counterpart, without any significant change in the magnitudes.
Lateral pressure profile
The lateral pressure profiles for all the systems simulated in this study are shown in Figs. 4 and 5. Specifically, Fig. 4 shows profiles for the bacterial lipid bilayer, together with the constituent POPE and POPG homogeneous systems. All three pressure profiles display a similar general trend. In particular, corresponding to the profile extremes at the outer aqueous phase, the lateral pressure is close to zero, as expected. Upon approaching the membrane, the lateral pressure rises and forms a positive peak at the interface between the water and lipid heads regions. Positive lateral pressure values indicate repulsive forces, acting to increase the area of the bilayer. Proceeding deeper into the bilayer, the lateral pressure declines sharply, forming deep troughs near the interface between the lipid heads and tails regions. Negative lateral pressure reflects the presence of attractive forces that act to reduce the bilayer area. In fact, the pressure troughs are a manifestation of the hydrophobic effect, whereby attractive lateral forces minimize the contact area between the hydrophobic core of the bilayer (comprising the hydrocarbon tails) and the hydrophilic environment (comprising lipid headgroups and water).
Lateral pressure profiles of different bilayer systems with varying compositions. Shaded areas represent error bars. The systems vary in complexity starting from the top, from pure, symmetric POPC a and PC:Chol b bilayers to the mammalian e and cancer f bilayers at the bottom. Pressure profiles from asymmetric bilayers c–f are plotted separately, with the ones on the right from the outer leaflet and the ones on the left from the inner leaflet
In the center of the bilayer, corresponding to the core of hydrophobic tails, the POPE and bacterial pressure profiles have five positive peaks, with four corresponding local minima in between them. POPG instead features a single central maximum. In general, positive pressure peaks in the lipid tails region represent repulsive forces arising from entropy loss [39]. Specifically, the tight molecular packing in the bilayer core forces the lipid tails to stretch, therefore losing entropy when compared to the isolated ”free” tails, ultimately causing significant intermolecular repulsion in the bilayer center [89, 90].
While all three profiles in Fig. 4 display similar qualitative features, important differences can be observed in relation to the different lipid composition. At the interface between the water and lipid heads regions, at a distance of approximately ∼2.5 nm from the bilayer center, the POPG system displays a maximum positive pressure of ∼530 bar, which is substantially larger than the value for the corresponding peaks in the bacterial (PE:PG) and POPE bilayers. Such large magnitude of the positive peak for POPG can be attributed to significant head–head electrostatic repulsion, a consequence of POPG’s net charge; in fact, we observed similarly high repulsion for a homogeneous bilayer comprising POPS, which also carries a net charge (see Fig. S4 in the Supplementary Information). Overall, in the water and heads regions, the bacterial bilayer displays a peak pressure of ∼400 bar, a value intermediate between the large POPG peak of ∼530 bar and the lower value of ∼140 bar for the pure POPE bilayer. The POPE pressure profile is also characterized by the presence of a small trough in the heads region at about ∼2.4 nm distance from the bilayer center, as previously observed for a DOPE bilayer [50]. Regarding the pressure troughs near the heads–tails interface, the POPE system features the most negative value of ∼-720 bar, while the bacterial and POPG bilayers feature pressures of ∼-550 and ∼-400 bar, respectively. In the tails region, the POPE and bacterial bilayers show large positive peaks in the range of ∼300 to ∼400 bar.
Regarding the mammalian and cancer bilayers, we have attempted to isolate the different contributions to the lateral pressure profiles from various lipid types. In particular, starting from the pure POPC bilayer, we have progressively added the required lipid species and monitored the corresponding effects on the pressure profile, until the attainment of the complete mammalian and cancer bilayer compositions. The results obtained for the various systems simulated are reported in Fig. 5. Starting from the top, the POPC bilayer (Fig. 5a) shows a pressure profile qualitatively similar to the previously discussed POPG bilayer, with large positive peaks in the heads region and a few small positive peaks in the tails region. The profile changes its structure and magnitude significantly with the addition of cholesterol (Fig. 5b); in particular, more peaks and troughs emerge, in agreement with previous studies [30, 31].
Noting that the cholesterol content in the mammalian bilayer is 34% of all lipids, we maintain constant cholesterol levels in both the upper and lower leaflets, while adding different lipids in different concentrations in the subsequent bilayers to make up the final lipid composition in the mammalian and cancer bilayers. The replacement of a proportion of POPC lipids with PSM (Fig. 5c) and PE (Fig. 5d) in the subsequent bilayers marginally increases the magnitudes of the peaks and troughs in the PC:Chol:PSM and PC:Chol:PSM:PE bilayers. The subsequent addition of PS lipids, which completes the formation of the mammalian bilayer, induces a small but noticeable reduction in the peak magnitudes (Fig. 5e). It can be observed that there is high similarity between the pressure profiles of PC:Chol and the more complex mammalian bilayers, indicating that cholesterol has a predominant role in shaping the lateral pressure profile. The PC:Chol and mammalian profiles are superimposed in Fig. S5 in the Supplementary Information. While the pressure profiles of the two bilayers are very similar in the outer leaflet, the mammalian pressure profile in the inner leaflet exhibits much higher magnitudes (up to ∼800 bar and ∼1070 bar at a distance of 1 nm and 1.75 nm from the bilayer center, respectively). The inner leaflet pressure profile of the mammalian bilayer also shows an outward shift in the lipid tails region, along with the appearance of a local trough in the lipid heads region (∼-175 bar at a distance of ∼2.7 nm from the bilayer center). This behavior could be attributed to a higher concentration of PE lipids (33%) in the inner leaflet of the mammalian bilayer, in qualitative agreement with our previous study on mixed DOPC/DOPE bilayers [50]. Compared to the mammalian system, the cancer bilayer (Fig. 5f) features greatly reduced peak magnitudes, which can be ascribed to the halving of its cholesterol content.
Dipole potential
The dipole potential profiles ψ(z) for POPG, POPE, and the mixed PE:PG bacterial systems are displayed in Fig. 6, while the profiles for the cancer and mammalian bilayers are displayed in Fig. 7. While all profiles share similar qualitative characteristics, there are significant composition-dependent differences in the magnitude of the potentials.
Figure 6 shows that, starting from the initial zero reference value in the aqueous phase, the dipole potential rises rapidly across the lipid heads region, up to peak values of ∼370 mV for POPG, ∼530 mV for the bacterial bilayer and ∼640 mV for POPE, at a distance of ∼1.3 nm from the bilayer center. The profiles then form a local minimum at ∼0.8 nm from the bilayer center, at a depth roughly corresponding to the presence of the lipid tails unsaturation. The profiles then reach a global maximum in the bilayer center, with values of ∼500 mV for POPG, ∼750 mV for the bacterial bilayer and ∼900 mV for the POPE bilayer. These central peaks are associated with the presence of partial charges, and corresponding dipoles, at the tails methyl termini. Our results for POPE are consistent with the previously reported atomistic simulation results of pure DOPC, DOPE, and mixed DOPC–DOPE bilayers [50, 70, 91] while for the POPG bilayers, our profile agrees with previous results by Zhao et al. [92].
Figure 7 reports the dipole potential profiles for the mammalian and cancer bilayers. Starting from the zero reference value in the water phase, the potential profiles rise towards the heads region up to peak values of ∼540 mV for the mammalian and ∼560 mV for the cancer system. Proceeding towards the bilayer center, a noticeable local minimum is present (although less prominent compared to the bacterial lipid bilayer results considered previously). The profiles then form a global maximum in the bilayer center, with a peak value of ∼985 mV for the mammalian and ∼920 mV for the cancer system. Upon entering the internal leaflet (corresponding to positive distances from the bilayer center in our convention), the profiles drop towards the secondary peaks near the heads–tails interface region, with values of ∼555 mV for the mammalian and ∼525 mV for the cancer system. Overall, these results show that the dipole potential is not significantly sensitive to changes in cholesterol and PS lipids.
Conclusions
In this work, we have used atomistic molecular dynamics simulations to study structural and depth-dependent properties of complex lipid bilayers with biologically relevant lipid compositions. In particular, we analyzed key properties of a model bacterial bilayer in terms of the contributions from the constituent lipid types. With regards to the lateral pressure profile, we found that the 2:1 POPE:POPG bacterial bilayer profile is mostly shaped by the POPE contribution in the central tails region, characterized by prominent pressure peaks of high magnitude, while the POPG contribution is predominant in the water and heads region. Regarding the dipole potential, the profiles of the bacterial system and those of the constituent lipids share the same qualitative features, while the magnitudes reflect the different composition. In particular, the bacterial profile, while bounded by the POPE and POPG bilayer profiles, has magnitudes closer to those of the POPE bilayer, consistently with the higher POPE concentration, compared to POPG, in the bacterial system. We also focused on the effects of changes in the lipid composition between mammalian and cancer bilayers. Cholesterol was identified as the predominant contributor to the lateral pressure in both complex bilayers. Specifically, the peak pressure magnitudes in the cancer profile were observed to be sharply reduced compared to those in the mammalian profile, corresponding to a 50% decrease in cholesterol in the cancer bilayer compared to the mammalian one. However, regarding the dipole potential, no noticeable differences were observed between the mammalian and cancer systems. To our knowledge, this is the first report of transmembrane lateral pressure and dipole potential profiles for model bacterial, mammalian, and cancer lipid bilayers.
References
Mouritsen OG (2005) Life - as a matter of fat. The emerging science of lipidomics, 1st edn. Springer, Berlin
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular biology of the cell, 4th edn. Garland Science
Smith AW (2012) Lipid–protein interactions in biological membranes: a dynamic perspective. Biochimica et Biophysica Acta (BBA) - Biomembranes 1818(2):172–177
Spector AA, Yorek MA (1985) Membrane lipid composition and cellular function. J Lipid Res 26 (9):1015–1035
Prausnitz MR, Langer R (2008) Transdermal drug delivery. Nat Biotechnol 26(11):1261–1268
Orsi M, Essex JW (2010) Permeability of drugs and hormones through a lipid bilayer: insights from dual-resolution molecular dynamics. Soft Matter 6(16):3797
Martin LJ, Chao R, Corry B (2014) Molecular dynamics simulation of the partitioning of benzocaine and phenytoin into a lipid bilayer. Biophys Chem 185:98–107
Lyubartsev AP, Rabinovich AL (2011) Recent development in computer simulations of lipid bilayers. Soft Matter 7(1):25–39
Orsi M, Sanderson W, Essex JW (2007) Coarse-grain modelling of lipid bilayers: a literature review. In: Hicks MG, Kettner C (eds) Molecular interactions - bringing chemistry to life. Beilstein-Institut, Frankfurt, pp 185–205
Tieleman DP, Marrink SJ, Berendsen HJC (1997) A computer perspective of membranes: molecular dynamics studies of lipid bilayer systems. Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes 1331 (3):235–270
Piggot TJ, Piñeiro Á, Khalid S (2012) Molecular dynamics simulations of phosphatidylcholine membranes: a comparative force field study. J Chem Theory Comput 8(11):4593–4609
Samuli Ollila OH, Vattulainen I (2010) Chapter 2 lateral pressure profiles in lipid membranes: dependence on molecular composition. In: Molecular simulations and biomembranes: from biophysics to function. The Royal Society of Chemistry, pp 26–55
Siani P, de Souza RM, Dias LG, Itri R, Khandelia H (2016) An overview of molecular dynamics simulations of oxidized lipid systems, with a comparison of ELBA and MARTINI force fields for coarse-grained lipid simulations. Biochimica et Biophysica Acta (BBA) - Biomembranes 1858(10):2498–2511
Grasso G, Muscat S, Rebella M, Morbiducci U, Audenino A, Danani A, Deriu MA (2018) Cell penetrating peptide modulation of membrane biomechanics by molecular dynamics. J Biomech 73:137–144
van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9(2):112–124
van Meer G, de Kroon AIPM (2011) Lipid map of the mammalian cell. J Cell Sci 124(Pt 1):5–8
Shevchenko A, Simons K (2010) Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11(8):593–598
Op den Kamp JAF (1979) Lipid asymmetry in membranes. Annu Rev Biochem 48(1):47–71
Gordesky SE, Marinetti GV (1973) The asymmetric arrangement of phospholipids in the human erythrocyte membrane. Biochem Biophys Res Commun 50(4):1027–1031
Verkleij AJ, Zwaal RF, Roelofsen B, Comfurius P, Kastelijn D, van Deenen LL (1973) The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochimica et Biophysica Acta (BBA) - Biomembranes 323(2): 178–193
Rothman JE, Lenard J (1977) Membrane asymmetry. Science 195(4280):743–753
Utsugi T, Schroit AJ, Connor J, Bucana CD, Fidler IJ (1991) Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res 51(11):3062–3066
Connor J, Bucana C, Fidler IJ, Schroit AJ (1989) Differentiation-dependent expression of phosphatidylserine in mammalian plasma membranes: quantitative assessment of outer-leaflet lipid by prothrombinase complex formation. Proc Natl Acad Sci U S A 86(9):3184–3188
Iwasaki T, Ishibashi J, Tanaka H, Sato M, Ai A, Taylor D, Yamakawa M (2009) Selective cancer cell cytotoxicity of enantiomeric 9-mer peptides derived from beetle defensins depends on negatively charged phosphatidylserine on the cell surface. Peptides 30(4):660–668
Hoskin DW, Ramamoorthy A (2008) Studies on anticancer activities of antimicrobial peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes 1778(2):357–375
Oldfield E, Meadows M, Rice D, Jacobs R (1978) Spectroscopic studies of specifically deuterium labeled membrane systems. Nuclear magnetic resonance investigation of the effects of cholesterol in model systems. Biochemistry 17(14):2727–2740
Urbina JA, Pekerar S, Le HB, Patterson J, Montez B, Oldfield E (1995) Molecular order and dynamics of phosphatidylcholine bilayer membranes in the presence of cholesterol, ergosterol and lanosterol: a comparative study using 2h-, 13C- and 31P-NMR spectroscopy. Biochimica et Biophysica Acta (BBA) - Biomembranes 1238(2):163–176
Thalia T, Mills G, Toombes ES, Tristram-Nagle S, Smilgies D-M, Feigenson GW, Nagle JF (2008) Order parameters and areas in fluid-phase oriented lipid membranes using wide angle X-ray scattering. Biophys J 95(2):669–681
Filippov A, Orädd G, Lindblom G (2003) The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J 84(5):3079–3086
Patra M (2005) Lateral pressure profiles in cholesterol-DPPC bilayers. Eur Biophys J 35(1):79–88
Ollila OHS, Rog T, Karttunen M, Vattulainen I (2007) Role of sterol type on lateral pressure profiles of lipid membranes affecting membrane protein functionality: comparison between cholesterol, desmosterol, 7-dehydrocholesterol and ketosterol. J Struct Biol 159(2 SPEC. ISS.):311–323
Klȧhn M, Zacharias M (2013) Transformations in plasma membranes of cancerous cells and resulting consequences for cation insertion studied with molecular dynamics. Phys Chem Chem Phys: PCCP 15(34):14427–41
Sohlenkamp C, Geiger O (2015) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 1:133–159
Merz C, Knoll W, Textor M, Reimhult E (2008) Formation of supported bacterial lipid membrane mimics. Biointerphases 3(2):41–50
Horn JN, Sengillo JD, Lin D, Romo TD, Grossfield A (2012) Characterization of a potent antimicrobial lipopeptide via coarse-grained molecular dynamics. Biochim Biophys Acta Biomembr 1818(2):212–218
Horn JN, Romo TD, Grossfield A (2013) Simulating the mechanism of antimicrobial lipopeptides with all-atom molecular dynamics. Biochemistry 52(33):5604–5610
Wang L (2012) Measurements and implications of the membrane dipole potential. Annu Rev Biochem 81:615–635
Cantor RS (1999) Lipid composition and the lateral pressure profile in bilayers. Biophys J 76(5):2625–2639
Cantor RS (1997) Lateral pressure in cell membranes: a mechanism for modulation of protein function. J Phys Chem 101(96):1723–1725
Cevc G, Marsh D (1987) Phospholipid bilayers: physical principles and models. Cell biology. Wiley
Templer RH, Castle SJ, Curran AR, Rumbles G, Klug DR (1998) Sensing isothermal changes in the lateral pressure in model membranes using di-pyrenyl phosphatidylcholine. Faraday Discuss 111:41–53. discussion 69–78
Kamo T, Handa T, Nakano M (2013) Lateral pressure change on phase transitions of phosphatidylcholine/diolein mixed membranes, vol 104
Orsi M, Noro MG, Essex JW (2011) Dual-resolution molecular dynamics simulation of antimicrobials in biomembranes. J R Soc Interface 6(59):826–841
Marsh D (2007) Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys J 93(11):3884–3899
van den Brink-van der Laan E, Killian JA, de Kruijff Ben (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochimica et Biophysica Acta (BBA) - Biomembranes 1666(1–2): 275–288
Cantor R S (1997) The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry 36(9):2339–2344
Ollila S, Hyvönen MT, Vattulainen I (2007) Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J Phys Chem B 111(12):3139–3150
Ding W, Palaiokostas M, Shahane G, Wang W, Orsi M (2017) Effects of high pressure on phospholipid bilayers. J Phys Chem B 121(41):9597–9606
Orsi M, Essex JW (2013) Physical properties of mixed bilayers containing lamellar and nonlamellar lipids: insights from coarse-grain molecular dynamics simulations. Faraday Discuss, 1–24
Ding W, Palaiokostas M, Wang W, Orsi M (2015) Effects of lipid composition on bilayer membranes quantified by All-Atom molecular dynamics. J Phys Chem B 119:15263–15274
Perrin SB, Sodt AJ, Cotten ML, Pastor RW (2014) The curvature induction of surface-bound antimicrobial peptides piscidin 1 and piscidin 3 varies with lipid chain length. J Membrane Biol 248(3):455–467
Clarke RJ (2001) The dipole potential of phospholipid membranes and methods for its detection. Adv Colloid Interface Sci 89-90:263–281
Liberman EA, Topaly VP (1969) Permeability of bimolecular phospholipid membranes for fat-soluble ions. Biofizika 14(3):452–461
Starke-Peterkovic T, Turner Nsl, Else PL, Clarke RJ (2005) Electric field strength of membrane lipids from vertebrate species: membrane lipid composition and Na+-K+-ATPase molecular activity. Am J Physiol Regul Integr Comp Physiol 288(3):63–70
Starke-Peterkovic T, Clarke RJ (2009) Effect of headgroup on the dipole potential of phospholipid vesicles. Eur Biophys J 39(1):103–110
Balleza D, Garcia-Arribas AB, Sot J, Ruiz-Mirazo K, Goni FM (2014) Ether- versus ester-linked phospholipid bilayers containing either linear or branched apolar chains. Biophys J 107(6):1364–1374
Orsi M, Michel J, Essex JW (2010) Coarse-grain modelling of DMPC and DOPC lipid bilayers. J Phys Condens Matter 22(15):155106
Horn JN, Romo TD, Grossfield A (2013) Simulating the mechanism of antimicrobial lipopeptides with all-atom molecular dynamics. Biochemistry 52(33):5604–5610
MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M (1998) All-Atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102(18):3586–3616
Klauda JB, Venable RM, Alfredo FJ, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr, Pastor RW (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114(23):7830–7843
Wu EL, Xi C, Jo S, Rui H, Song KC, Dávila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, Im W (2014) CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem 35(27): 1997–2004
Jo S, Lim JB, Klauda JB, Im W (2009) CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J 97(1):50–58
Jo S, Kim T, Im W (2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2(9):880
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a Web-based graphical user interface for CHARMM. J Comput Chem 29(11):1859–1865
Doktorova M, Weinstein H (2018) Accurate in silico modeling of asymmetric bilayers based on biophysical principles. Biophys J 115(9):1638–1643
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ C (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126(1):014101
Venable RM, Brown FLH, Pastor RW (2015) Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem Phys Lipids 192:60–74
Siu SWI, Vácha R, Jungwirth P, Böckmann RA (2008) Biomolecular simulations of membranes: physical properties from different force fields, vol 128
Shinoda K, Shinoda W, Baba T, Mikami M (2004) Comparative molecular dynamics study of ether- and ester-linked phospholipid bilayers. J Chem Phys 121(19):9648–9654
Orsi M (2014) Comparative assessment of the ELBA coarse-grained model for water. Mol Phys 112:1566–1576
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190
Shuichi M, Kollman PA (1992) Settle: an analytical version of the shake and rattle algorithm for rigid water models. J Comput Chem 13(8):952–962
Berk H, Henk B, Herman BJC, Johannes FGEM (1997) Lincs: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Lukat G, Krüger J, Sommer B (2013) APL@Voro: a Voronoi-based membrane analysis tool for GROMACS trajectories. J Chem Inf Model 53(11):2908–2925
Vanegas JM, Torres-Sánchez A, Arroyo M (2014) Importance of force decomposition for local stress calculations in biomembrane molecular simulations. J Chem Theory Comput 10(2):691– 702
Torres-Sánchez A, Vanegas JM, Arroyo M (2015) Examining the mechanical equilibrium of microscopic stresses in molecular simulations. Phys Rev Lett 114(25):258102
Samuli Ollila OH, Jelger Risselada H, Louhivuori M, Lindahl E, Vattulainen I, Marrink SJ (2009) 3D pressure field in lipid membranes and membrane–protein complexes. Phys Rev Lett 102(7):078101
Thijsse BJ, Hollanders MA, Hendrikse J (1998) A practical algorithm for least-squares spline approximation of data containing noise. Comput Phys 12(4)
McIntosh TJ (1978) The effect of cholesterol on the structure of phosphatidylcholine bilayers. Biochimica et Biophysica Acta (BBA) - Biomembranes 513(1):43–58
Hofsäss C, Lindahl E, Edholm O (2003) Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys J 84(4):2192–2206
Leftin A, Molugu TR, Job C, Beyer K, Brown MF (2014) Area per lipid and cholesterol interactions in membranes from separated local-field 13C NMR spectroscopy. Biophys J 107(10):2274–2286
de Meyer F, Smit B (2009) Effect of cholesterol on the structure of a phospholipid bilayer. Proc Natl Acad Sci USA 106(10):3654–3658
Kučerka N, Pencer J, Nieh M-P, Katsaras J (2007) Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur Phys J E Soft Matter 23(3):247–254
Asawakarn T, Cladera J, O’Shea P (2001) Effects of the membrane dipole potential on the interaction of saquinavir with phospholipid membranes and plasma membrane receptors of Caco-2 cells. J Biol Chem 276 (42):38457–38463
Lipowsky R, Sackmann E (1995) Structure and dynamics of membranes. Number v. 1, pt. 1 in Handbook of biological physics. Elsevier Science
Mukhin SI, Baoukina S (2005) Analytical derivation of thermodynamic characteristics of lipid bilayer from a flexible string model. Phys Rev E 71(6):061918
Srinivas Reddy A, Warshaviak DT, Chachisvilis M (2012) Effect of membrane tension on the physical properties of DOPC lipid bilayer membrane. Biochim Biophys Acta Biomembr 1818(9):2271–2281
Zhao W, Rȯg T, Gurtovenko AA, Vattulainen I, Karttunen M (2007) Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidylglycerol lipid bilayers with Na+ counterions. Biophys J 92 (4):1114–24
Kuvcerka N, Nieh M-P, Katsaras J (2011) Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochimica et Biophysica Acta (BBA) - Biomembranes 1808(11):2761–2771
Kucerka N, Tristram-Nagle S, Nagle JF (2005) Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol 208(3):193–202
Skjevik ÅA, Madej BD, Dickson CJ, Teigen K, Walker RC, Gould IR (2015) All-atom lipid bilayer self-assembly with the AMBER and CHARMM lipid force fields. Chem Commun 51(21):4402–4405
Rand RP, Fuller N, Parsegian VA, Rau DC (1988) Variation in hydration forces between neutral phospholipid bilayers: evidence for hydration attraction. Biochemistry 27(20):7711–7722
Rappolt M, Hickel A, Bringezu F, Lohner K (2003) Mechanism of the lamellar/inverse hexagonal phase transition examined by high-resolution X-ray diffraction. Biophys J 84(5):3111– 3122
Kuvčerka N, Holland BW, Gray CG, Tomberli B, Katsaras J (2012) Scattering density profile model of POPG bilayers as determined by molecular dynamics simulations and small-angle neutron and X-ray scattering experiments. J Phys Chem B 116(1):232–239
Tolokh IS, Vivcharuk V, Tomberli B, Gray CG (2009) Binding free energy and counterion release for adsorption of the antimicrobial peptide lactoferricin B on a POPG membrane. Phys Rev E 80(3):031911
Pan J, Heberle FA, Tristram-Nagle S, Szymanski M, Koepfinger M, Katsaras J, Kučerka N (2012) Molecular structures of fluid phase phosphatidylglycerol bilayers as determined by small-angle neutron and X-ray scattering. Biochimica et Biophysica Acta (BBA) - Biomembranes 1818(9):2135–2148
Skjevik ÅA, Madej BD, Dickson CJ, Lin C, Teigen K, Walker RC, Gould IR (2016) Simulation of lipid bilayer self-assembly using all-atom lipid force fields. Phys Chem Chem Phys: PCCP 18(15):10573–10584
Pan J, Cheng X, Monticelli L, Heberle FA, Kučerka N, Tieleman DP, Katsaras J (2014) The molecular structure of a phosphatidylserine bilayer determined by scattering and molecular dynamics simulations. Soft Matter 10(21):3716–3725
Mukhopadhyay P, Monticelli L, Tieleman DP (2004) Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with na+ counterions and NaCl. Biophys J 86(3):1601–1609
Olsen BN, Bielska AA, Lee T, Daily MD, Covey DF, Schlesinger PH, Baker NA, Ory DS (2013) The structural basis of cholesterol accessibility in membranes. Biophys J 105(8):1838–1847
Loura LMS, do Canto AMTM, Martins J (2013) Sensing hydration and behavior of pyrene in POPC and POPC/cholesterol bilayers: a molecular dynamics study. Biochimica et Biophysica Acta (BBA) - Biomembranes 1828(3):1094–1101
Hong C, Peter Tieleman D, Yi W (2014) Microsecond molecular dynamics simulations of lipid mixing. Langmuir 30(40):11993–12001
Murzyn K, Róg T (2005) Marta Pasenkiewicz-Gierula phosphatidylethanolamine-phosphatidylglycerol bilayer as a model of the inner bacterial membrane. Biophys J 88(2): 1091–1103
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–8
Acknowledgments
This project made use of time on the ARCHER supercomputer via the UK High-End Computing Consortium for Biomolecular Simulation (HECBioSim), supported by EPSRC (grant no. EP/L000253/1). GS acknowledges financial support from the Institute of Bioengineering at Queen Mary University of London.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shahane, G., Ding, W., Palaiokostas, M. et al. Physical properties of model biological lipid bilayers: insights from all-atom molecular dynamics simulations. J Mol Model 25, 76 (2019). https://doi.org/10.1007/s00894-019-3964-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-019-3964-0