Abstract
Odorant binding proteins (OBPs) are important in insect olfactory recognition. These proteins bind specifically to insect semiochemicals and induce their seeking, mating, and alarm behaviors. Molecular docking and molecular dynamics simulations were performed to provide computational insight into the interaction mode between AgamOBP7 and novel (E)-β-farnesene (EBF) analogues with an aromatic ring. The ligand-binding cavity in OBP7 was found to be mostly hydrophobic due to the presence of several nonpolar residues. The interactions between the EBF analogues and the hydrophobic residues in the binding cavity increased in strength as the distance between them decreased. The EBF analogues with an N-methyl formamide or ester linkage had higher docking scores than those with an amide linkage. Moreover, delocalized π–π and electrostatic interactions were found to contribute significantly to the binding between the ligand benzene ring and nearby protein residues. To design new compounds with higher activity, four EBF analogues D1–D4 with a benzene ring were synthesized and evaluated based on their docking scores and binding affinities. D2, which had an N-methyl formamide group linkage, exhibited stronger binding than D1, which had an amide linkage. D4 exhibited particularly strong binding due to multiple hydrophobic interactions with the protein. This study provides crucial foundations for designing novel EBF analogues based on the OBP structure.

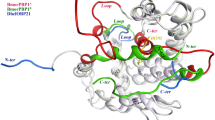
The design strategy of new EBF analogues based on the OBP7 structure






Similar content being viewed by others
References
Harris KF, Maramorosch K (1977) Aphids as virus vectors. Academic, New York, 4:435–454
Bowers WS, Nault LR, Webb RE, Dutky SR (1972) Aphid alarm pheromone: isolation, identification, synthesis. Science 177:1121–1122
Dawson GW, Giffiths DC, Pickett JA, Plumb RT, Woodcock CM, Zhang ZN (1988) Structure–activity studies on aphid alarm pheromone derivatives and their field use against transmission of barley yellow dwarf virus. Pestic Sci 22:17–30
Li ZM, Wang TS, Yao EY (1987) Researches on insect pheromone III studies on aphid alarm pheromone mimics. Acta Chem Sin 45:1124–1128
Zhang ZN, Liu X, Pickett JA (1988) Several aphids alarm pheromone analogues possessing biological activity. Acta Entomol Sin 31:435–438
Sun L, Ling Y, Wang C, Sun YF, Rui CH, Yang XL (2011) Synthesis and biological activity of E-β-farnesene analogues containing substituent nitroguanidine. Chin J Org Chem 31:2061–2066
Kang TN, Ling Y, Rui CH, Yang XL, Fan XL, Chen FH (2008) Synthesis of E-β-farnesene analogues containing five-membered azaheterocycles and their biological activity. Chin J Org Chem 28:617–621
Sun YF, Li YQ, Ling Y, Yu HL, Yang SX, Yang XL (2011) Design, synthesis and biological activity of E-β-farnesene analogues containing pyrazole-carboxamide. Chin J Org Chem 31:1425–1432
Sun L (2013) Synthesis process and structure optimization of EBF analogue CAU1204. PhD dissertation. China Agricultural University, Beijing
Liu SH (2014) Design, synthesis and bioactivity of neonicotinoids and EBF analogues. PhD dissertation. China Agricultural University, Beijing
Pelosi P, Zhou JJ, Ban LP, Calvello M (2006) Soluble proteins in insect chemical communication. Cell Mol Life Sci 63:1658–1676
Tegoni M, Campanacci V, Cambillau C (2004) Structural aspects of sexual attraction and chemical communication in insects. Trends Biochem Sci 29:257–264
Qiao HL, Tuccori E, He XL, Gazzano A, Field L, Zhou JJ, Pelosi P (2009) Discrimination of alarm pheromone (E)-β-farnesene by aphid odorant-binding proteins. Insect Biochem Mol 39:414–419
Zhong T, Yin J, Deng SS, Li KB, Cao YZ (2012) Fluorescence competition assay for the assessment of green leaf volatiles and trans-β-farnesene bound to three odorant-binding proteins in the wheat aphid Sitobion avenae (Fabricius). J Insect Physiol 58:771–781
Sun YF, Qiao HL, Ling Y, Yang SX, Rui CH, Pelosi P, Yang XL (2011) New analogues of (E)-β-farnesene with insecticidal activity and binding affinity to aphid odorant-binding proteins. J Agric Food Chem 59:2456–2461
Tsitsanou KE, Thireou T, Drakou CE, Koussis K, Keramioti MV, Leonidas DD, Eliopoulos E, Iatrou K, Zographos SE (2012) Anopheles gambiae odorant binding protein crystal complex with the synthetic repellent DEET: implications for structure-based design of novel mosquito repellents. Cell Mol Life Sci 69:283–297
Mao Y, Xu XZ, Xu W, Ishida Y, Leal WS, Ames JB, Clardy J (2010) Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci USA 107:19102–19107
Leite NR, Krogh R, Xu W, Ishida Y, Iulek J, Leal WS, Oliva G (2009) Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive “lid”. PLoS ONE 4:e8006
Lagarde A, Spinelli S, Tegoni M, He XL, Field L, Zhou JJ, Cambillau C (2011) The crystal structure of ordorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J Mol Biol 414:401–412
Davrazou F, Dong E, Murphy EJ, Johnson HT, Jones DNM (2011) New insights into the mechanism of odorant detection by the malaria-transmitting mosquito Anopheles gambiae. J Biol Chem 286:34175–34183
Spinelli S, Lagarde A, Iovinella I, Legrand P, Tegoni M, Pelosi P, Cambillau C (2012) Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem Mol Biol 42:41–50
Sun YF, De Biasio F, Qiao HL, Iovinella I, Yang SX, Ling Y, Riviello L, Battaglia D, Falabella P, Yang XL, Pelosi P (2012) Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-β-farnesene and structural analogues. PLoS ONE 7(3):e32759
Sun YF (2011) Discovery of novel aphid control agents based on OBPs: design, synthesis and bioactivity of EBF analogues. PhD dissertation. China Agricultural University, Beijing
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Schüttelkopf AW, van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D 60:1355–1363
Hess B, Bekker H, Berendsen HJC, Fraajie JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Essman U, Perela L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald potential. J Chem Phys 103:8577–8592
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian03, revision D.01. Gaussian, Inc., Wallingford
Jain AN (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46:499–511
Jain AN (1996) Scoring noncovalent protein–ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des 10:427–440
Kellenberger E, Rodrigo J, Muller P, Rognan D (2004) Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 57:225–242
Tripos Associates (2006) Sybyl version 7.3. Tripos Associates, St. Louis
Yi X, Zhang YB, Wang PD, Qi JW, Hu MY, Zhong GH (2015) Ligands binding and molecular simulation: the potential investigation of a biosensor based on an insect odorant binding protein. Int J Biol Sci 11:75–87
Salonen LM, Ellermann M, Diederich F (2011) Aromatic rings in chemical and biological recognition: energetics and structures. Angew Chem Int Ed 50:4808–4842
Janiak C (2000) A critical account on π–π stacking in metal complexes with aromatic nitrogen-containing ligands. J Chem Soc Dalton Trans 21:3885–389
Acknowledgments
We thank Dr. Pelosi for fruitful discussions and writing assistance. This work was supported by the National Scientific and Technology Supporting Program of China (2011BAE06B05-5) and the National Natural Science Foundation of China (21132003, 31371946).
Author contributions
S.S. Wang, S.Q. Du, and H.X. Duan conceived and designed the experiments. Y.F. Sun, Y.G. Qin, and X.L. Yang prepared the EBF analogues. S.S. Wang, Y.F. Sun, and H.X. Duan analyzed the data. S.S. Wang and H.X. Duan wrote the first draft of the manuscript. H.X. Duan and X.L. Yang made critical revisions and approved the final manuscript. All the authors reviewed and approved the final manuscript. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, S., Sun, Y., Du, S. et al. Computer-aided rational design of novel EBF analogues with an aromatic ring. J Mol Model 22, 144 (2016). https://doi.org/10.1007/s00894-016-3011-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-016-3011-3