Abstract
Objectives
The present systematic review aims to report and critically assess the findings of the available scientific evidence from genetic association studies examining the genetic variants underlying skeletal class III malocclusion and its sub-phenotypes.
Material and methods
A pre-piloted protocol was registered and followed. The PubMed, Scopus, WOS, Cochrane Library, Gray Open literature, and CADTH databases were explored for genetic association studies following PICOS-based selection criteria. The research was reported in accordance with PRISMA statement and HuGE guidelines. The Q-genie tool was applied to assess the quality of genetic studies. Meta-analysis of genetic association studies was done by means of Meta-Genyo tool.
Results
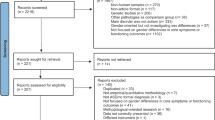
A total of 8258 articles were retrieved, of which 22 were selected for in-depth analysis. Most of the studies did not differentiate between sub-phenotypes, and the cohorts were heterogeneous regarding ethnicity. Four to five principal components of class III malocclusion explained the phenotypic variation, and gene variants at MYO1H(rs10850110), BMP3(rs1390319), GHR (rs2973015,rs6184, rs2973015), FGF7(rs372127537), FGF10(rs593307), and SNAI3(rs4287555) (p < .05) explained most of the variation across the studies, associated to vertical, horizontal, or combined skeletal discrepancies. Meta-analysis results identified a statistically significant association between risk of class III malocclusion of A allele of the FBN3 rs7351083 [OR 2.13; 95% CI 1.1–4.1; p 0.02; recessive model].
Conclusion
Skeletal class III is a polygenic trait substantially modulated by ethnicity. A multicentric approach should be considered in future studies to increase sample sizes, applying multivariate analysis such as PCA and cluster analysis to characterize existing sub-phenotypes warranting a deeper analysis of genetic variants contributing to skeletal class III craniofacial disharmony.
Clinical relevance
Grasping the underlying mechanisms of this pathology is critical for a fuller understanding of its etiology, allowing generation of preventive strategies, new individualized therapeutic approaches and more accurate treatment planification strategies.



Similar content being viewed by others
References
Nikopensius T, Saag M, Jagomägi T, Annilo T, Kals M, Kivistik PA, Milani L, Metspalu A (2013) A missense mutation in DUSP6 is associated with Class III malocclusion. J Dent Res 92(10):893–898. https://doi.org/10.1177/0022034513502790
Doraczynska-Kowalik A, Nelke KH, Pawlak W, Sasiadek MM, Gerber H (2017) Genetic Factors Involved in Mandibular Prognathism. J Craniofac Surg 28(5):e422–e431. https://doi.org/10.1097/scs.0000000000003627
de Frutos-Valle L, Martin C, Alarcon JA, Palma-Fernandez JC, Iglesias-Linares A (2019) Subclustering in Skeletal Class III Phenotypes of Different Ethnic Origins: A Systematic Review. J Evid Based Dent Pract 19(1):34–52. https://doi.org/10.1016/j.jebdp.2018.09.002
Kajii TS, Oka A, Saito F, Mitsui J, Iida J (2019) Whole-exome sequencing in a Japanese pedigree implicates a rare non-synonymous single-nucleotide variant in BEST3 as a candidate for mandibular prognathism. Bone 122:193–198. https://doi.org/10.1016/j.bone.2019.03.004
Liu H, Wu C, Lin J, Shao J, Chen Q, Luo E (2017) Genetic Etiology in Nonsyndromic Mandibular Prognathism. J Craniofac Surg 28(1):161–169. https://doi.org/10.1097/scs.0000000000003287
Sidlaukas Antanas LK (2009) The prevalence of malocclusion among 7–15-year-old Lithuanian schoolchildren. Medicina (Kaunas) 45(2):147–152
Perillo L, Masucci C, Ferro F, Apicella D, Baccetti T (2010) Prevalence of orthodontic treatment need in southern Italian schoolchildren. Eur J Orthod 32(1):49–53. https://doi.org/10.1093/ejo/cjp050
Nowrin S, Alam M, Basri R (2015) Genetic effect and prevalence of class iii malocclusion in different population: An overview. Int J Pharm Bio Sci 6:910–918
Moreno Uribe LM, Miller SF (2015) Genetics of the dentofacial variation in human malocclusion. Orthod Craniofacial Res 18(Suppl 1(0 1)):91–99. https://doi.org/10.1111/ocr.12083
da Fontoura CS, Miller SF, Wehby GL, Amendt BA, Holton NE, Southard TE, Allareddy V, Moreno Uribe LM (2015) Candidate Gene Analyses of Skeletal Variation in Malocclusion. J Dent Res 94(7):913–920. https://doi.org/10.1177/0022034515581643
Moreno Uribe LM, Ray A, Blanchette DR, Dawson DV, Southard TE (2015) Phenotypegenotype correlations of facial width and height proportions in patients with Class II malocclusion. Orthod Craniofacial Res 18(Suppl 1(0 1)):100–108. https://doi.org/10.1111/ocr.12084
Moreno Uribe LM, Vela KC, Kummet C, Dawson DV, Southard TE (2013) Phenotypic diversity in white adults with moderate to severe Class III malocclusion. Am J Orthod Dentofac Orthop 144(1):32–42. https://doi.org/10.1016/j.ajodo.2013.02.019
Claes P, Roosenboom J, White JD, Swigut T, Sero D, Li J, Lee MK, Zaidi A, Mattern BC, Liebowitz C, Pearson L, Gonzalez T, Leslie EJ, Carlson JC, Orlova E, Suetens P, Vandermeulen D, Feingold E, Marazita ML, Shaffer JR, Wysocka J, Shriver MD, Weinberg SM (2018) Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat Genet 50(3):414–423. https://doi.org/10.1038/s41588-018-0057-4
Moreno Uribe LM, Ray A, Blanchette DR, Dawson DV, Southard TE (2015) Phenotypegenotype correlations of facial width and height proportions in patients with Class II malocclusion. Orthod Craniofacial Res 18(Suppl 1):100–108. https://doi.org/10.1111/ocr.12084
Yamaguchi T, Park SB, Narita A, Maki K, Inoue I (2005) Genome-wide linkage analysis of mandibular prognathism in Korean and Japanese patients. J Dent Res 84(3):255–259. https://doi.org/10.1177/154405910508400309
Cruz CV, Mattos CT, Maia JC, Granjeiro JM, Reis MF, Mucha JN, Vilella B, Ruellas AC, Luiz RR, Costa MC, Vieira AR (2017) Genetic polymorphisms underlying the skeletal Class III phenotype. Am J Orthod Dentofac Orthop 151(4):700–707. https://doi.org/10.1016/j.ajodo.2016.09.013
Cruz RM, Krieger H, Ferreira R, Mah J, Hartsfield J Jr, Oliveira S (2008) Major gene and multifactorial inheritance of mandibular prognathism. Am J Med Genet A 146a(1):71–77. https://doi.org/10.1002/ajmg.a.32062
Jang JY, Park EK, Ryoo HM, Shin HI, Kim TH, Jang JS, Park HS, Choi JY, Kwon TG (2010) Polymorphisms in the Matrilin-1 gene and risk of mandibular prognathism in Koreans. J Dent Res 89(11):1203–1207. https://doi.org/10.1177/0022034510375962
Xue F, Wong R, Rabie AB (2010) Identification of SNP markers on 1p36 and association analysis of EPB41 with mandibular prognathism in a Chinese population. Arch Oral Biol 55(11):867–872. https://doi.org/10.1016/j.archoralbio.2010.07.018
Ikuno K, Kajii TS, Oka A, Inoko H, Ishikawa H, Iida J (2014) Microsatellite genome-wide association study for mandibular prognathism. Am J Orthod Dentofac Orthop 145(6):757–762. https://doi.org/10.1016/j.ajodo.2014.01.022
Saito F, Kajii TS, Oka A, Ikuno K, Iida J (2017) Genome-wide association study for mandibular prognathism using microsatellite and pooled DNA method. Am J Orthod Dentofac Orthop 152(3):382–388. https://doi.org/10.1016/j.ajodo.2017.01.021
Frazier-Bowers S, Rincon-Rodriguez R, Zhou J, Alexander K, Lange E (2009) Evidence of linkage in a Hispanic cohort with a Class III dentofacial phenotype. J Dent Res 88(1):56–60. https://doi.org/10.1177/0022034508327817
Cunha A, Nelson-Filho P, Maranon-Vasquez GA, Ramos AGC, Dantas B, Sebastiani AM, Silverio F, Omori MA, Rodrigues AS, Teixeira EC, Levy SC, Araujo MC, Matsumoto MAN, Romano FL, Antunes LAA, Costa DJD, Scariot R, Antunes LS, Vieira AR, Kuchler EC (2019) Genetic variants in ACTN3 and MYO1H are associated with sagittal and vertical craniofacial skeletal patterns. Arch Oral Biol 97:85–90. https://doi.org/10.1016/j.archoralbio.2018.09.018
Jheon AH, Oberoi S, Solem RC, Kapila S (2017) Moving towards precision orthodontics: An evolving paradigm shift in the planning and delivery of customized orthodontic therapy. Orthod Craniofacial Res 20(Suppl 1):106–113. https://doi.org/10.1111/ocr.12171
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6(7):e1000097. https://doi.org/10.1371/journal.pmed.1000097
Little JHJe (2006) The HuGENetTM HuGE Review Handbook, version 1.0.
Saleh AA, Ratajeski MA, Bertolet M (2014) Grey Literature Searching for Health Sciences Systematic Reviews: A Prospective Study of Time Spent and Resources Utilized. Evid Based Libr Inf Pract 9(3):28–50. https://doi.org/10.18438/b8dw3k
(EPOC) CEPaOoC (2017) How to develop a search strategy. EPOC Resources for review authors. http://epoc.cochrane.org/resources/epoc-resources-review-authors
Sohani ZN, Meyre D, de Souza RJ, Joseph PG, Gandhi M, Dennis BB, Norman G, Anand SS (2015) Assessing the quality of published genetic association studies in meta-analyses: the quality of genetic studies (Q-Genie) tool. BMC Genet 16:50. https://doi.org/10.1186/s12863-015-0211-2
Martorell-Marugan J, Toro-Dominguez D, Alarcon-Riquelme ME, Carmona-Saez P (2017) MetaGenyo: a web tool for meta-analysis of genetic association studies. BMC Bioinformatics 18(1):563. https://doi.org/10.1186/s12859-017-1990-4
Nakaoka H, Inoue I (2009) Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner's curse. J Hum Genet 54(11):615–623. https://doi.org/10.1038/jhg.2009.95
Rodrigues AS, Teixeira EC, Antunes LS, Nelson-Filho P, Cunha AS, Levy SC, de Souza Araújo MT, de Carvalho Ramos AG, Cruz GV, Omori MA, Matsumoto MAN, Vieira AR, Küchler EC, Marañón-Vásquez GA, Antunes LAA (2020) Association between craniofacial morphological patterns and tooth agenesis-related genes. Prog Orthod 21(1):9. https://doi.org/10.1186/s40510-020-00309-5
Marañón-Vásquez GA, Vieira AR, de Carvalho Ramos AG, Dantas B, Romano FL, Palma-Dibb RG, Arid J, Carpio K, Nelson-Filho P, de Rossi A, Scariot R, Levy SC, Antunes LAA, Antunes LS, Küchler EC (2020) GHR and IGF2R genes may contribute to normal variations in craniofacial dimensions: Insights from an admixed population. Am J Orthod Dentofac Orthop 158(5):722–730.e716. https://doi.org/10.1016/j.ajodo.2019.10.020
Marañón-Vásquez GA, Dantas B, Kirschneck C, Arid J, Cunha A, Ramos AGC, Omori MA, Rodrigues AS, Teixeira EC, Levy SC, Schroeder A, Matsumoto MAN, Proff P, Antunes LAA, Vieira AR, Antunes LS, Küchler EC (2019) Tooth agenesis-related GLI2 and GLI3 genes may contribute to craniofacial skeletal morphology in humans. Arch Oral Biol 103:12–18. https://doi.org/10.1016/j.archoralbio.2019.05.008
Guan X, Song Y, Ott J, Zhang Y, Li C, Xin T, Li Z, Gan Y, Li J, Zhou S, Zhou Y (2015) The ADAMTS1 Gene Is Associated with Familial Mandibular Prognathism. J Dent Res 94(9):1196–1201. https://doi.org/10.1177/0022034515589957
Jiang Q, Zhou X, Cheng L, Li M (2019) The Adhesion and Invasion Mechanisms of Streptococci. Curr Issues Mol Biol 32:521–560. https://doi.org/10.21775/cimb.032.521
Xue F, Rabie AB, Luo G (2014) Analysis of the association of COL2A1 and IGF-1 with mandibular prognathism in a Chinese population. Orthod Craniofacial Res 17(3):144–149. https://doi.org/10.1111/ocr.12038
Xiong X, Li S, Cai Y, Chen F (2017) Targeted sequencing in FGF/FGFR genes and association analysis of variants for mandibular prognathism. Medicine (Baltimore) 96(25):e7240. https://doi.org/10.1097/md.0000000000007240
Sun R, Wang Y, Jin M, Chen L, Cao Y, Chen F (2018) Identification and Functional Studies of MYO1H for Mandibular Prognathism. J Dent Res 97(13):1501–1509. https://doi.org/10.1177/0022034518784936
Tobón-Arroyave SI, Jiménez-Arbeláez GA, Alvarado-Gómez VA, Isaza-Guzmán DM, Flórez-Moreno GA, Pérez-Cano MI (2018) Association analysis between rs6184 and rs6180 polymorphisms of growth hormone receptor gene regarding skeletal-facial profile in a Colombian population. Eur J Orthod 40(4):378–386. https://doi.org/10.1093/ejo/cjx070
Sasaki Y, Satoh K, Hayasaki H, Fukumoto S, Fujiwara T, Nonaka K (2009) The P561T polymorphism of the growth hormone receptor gene has an inhibitory effect on mandibular growth in young children. Eur J Orthod 31(5):536–541. https://doi.org/10.1093/ejo/cjp017
Gupta P, Chaturvedi TP, Sharma V (2017) Expressional Analysis of MSX1 (Human) Revealed its Role in Sagittal Jaw Relationship. J Clin Diagn Res 11(8):Zc71–zc77. https://doi.org/10.7860/jcdr/2017/26755.10441
Ghergie M, Feştila D, Lupan I, Popescu O, Kelemen BS (2013) Testing the association between orthodonthic classes I, II, III and SNPs (rs731236, rs8004560, rs731236) in a Romanian clinical sample. Ann Romanian Soc Cell Biol 18:43–51
Bayram S, Basciftci FA, Kurar E (2014) Relationship between P561T and C422F polymorphisms in growth hormone receptor gene and mandibular prognathism. Angle Orthod 84(5):803–809. https://doi.org/10.2319/091713-680.1
Weaver CA, Miller SF, da Fontoura CS, Wehby GL, Amendt BA, Holton NE, Allareddy V, Southard TE, Moreno Uribe LM (2017) Candidate gene analyses of 3-dimensional dentoalveolar phenotypes in subjects with malocclusion. Am J Orthod Dentofac Orthop 151(3):539–558. https://doi.org/10.1016/j.ajodo.2016.08.027
Tassopoulou-Fishell M, Deeley K, Harvey EM, Sciote J, Vieira AR (2012) Genetic variation in myosin 1H contributes to mandibular prognathism. Am J Orthod Dentofac Orthop 141(1):51–59. https://doi.org/10.1016/j.ajodo.2011.06.033
Klingenberg CP (2016) Size, shape, and form: concepts of allometry in geometric morphometrics. Dev Genes Evol 226(3):113–137. https://doi.org/10.1007/s00427-016-0539-2
Signorelli L, Patcas R, Peltomaki T, Schatzle M (2016) Radiation dose of cone-beam computed tomography compared to conventional radiographs in orthodontics. J Orofac Orthop 77(1):9–15. https://doi.org/10.1007/s00056-015-0002-4
Huang D, Petrykowska HM, Miller BF, Elnitski L, Ovcharenko I (2019) Identification of human silencers by correlating cross-tissue epigenetic profiles and gene expression. Genome Res 29(4):657–667. https://doi.org/10.1101/gr.247007.118
Sun YV, Hu YJ (2016) Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv Genet 93:147–190. https://doi.org/10.1016/bs.adgen.2015.11.004
Wafi A, Mirnezami R (2018) Translational -omics: Future potential and current challenges in precision medicine. Methods 151:3–11. https://doi.org/10.1016/j.ymeth.2018.05.009
UniProt C (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47(D1):D506–D515. https://doi.org/10.1093/nar/gky1049
Arun RM, Lakkakula BV, Chitharanjan AB (2016) Role of myosin 1H gene polymorphisms in mandibular retrognathism. Am J Orthod Dentofac Orthop 149(5):699–704. https://doi.org/10.1016/j.ajodo.2015.10.028
Sella-Tunis T, Pokhojaev A, Sarig R, O'Higgins P, May H (2018) Human mandibular shape is associated with masticatory muscle force. Sci Rep 8(1):6042. https://doi.org/10.1038/s41598-018-24293-3
Wieczorek A, Loster J, Loster BW (2013) Relationship between occlusal force distribution and the activity of masseter and anterior temporalis muscles in asymptomatic young adults. Biomed Res Int 2013:354017. https://doi.org/10.1155/2013/354017
Iodice G, Danzi G, Cimino R, Paduano S, Michelotti A (2016) Association between posterior crossbite, skeletal, and muscle asymmetry: a systematic review. Eur J Orthod 38(6):638–651. https://doi.org/10.1093/ejo/cjw003
Qi K, Guo SX, Xu Y, Deng Q, Liu L, Li B, Wang MQ (2016) An investigation of the simultaneously recorded occlusal contact and surface electromyographic activity of jawclosing muscles for patients with temporomandibular disorders and a scissors-bite relationship. J Electromyogr Kinesiol 28:114–122. https://doi.org/10.1016/j.jelekin.2016.04.004
Toro-Ibacache V, Ugarte F, Morales C, Eyquem A, Aguilera J, Astudillo W (2019) Dental malocclusions are not just about small and weak bones: assessing the morphology of the mandible with cross-section analysis and geometric morphometrics. Clin Oral Investig 23(9):3479–3490. https://doi.org/10.1007/s00784-018-2766-6
Lu Y, Li Y, Cavender AC, Wang S, Mansukhani A, D'Souza RN (2012) Molecular studies on the roles of Runx2 and Twist1 in regulating FGF signaling. Dev Dyn 241(11):1708–1715. https://doi.org/10.1002/dvdy.23858
Lee SK, Kim YS, Oh HS, Yang KH, Kim EC, Chi JG (2001) Prenatal development of the human mandible. Anat Rec 263(3):314–325. https://doi.org/10.1002/ar.1110
Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, Faden DL, Rimbault M, Decker B, Kidd JM, Sood R, Boyko AR, Fondon JW 3rd, Wayne RK, Bustamante CD, Ciruna B, Ostrander EA (2012) Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet 8(8):e1002849. https://doi.org/10.1371/journal.pgen.1002849
Fu X, Xu J, Chaturvedi P, Liu H, Jiang R, Lan Y (2017) Identification of Osr2 Transcriptional Target Genes in Palate Development. J Dent Res 96(12):1451–1458. https://doi.org/10.1177/0022034517719749
Perillo L, Monsurro A, Bonci E, Torella A, Mutarelli M, Nigro V (2015) Genetic association of ARHGAP21 gene variant with mandibular prognathism. J Dent Res 94(4):569–576. https://doi.org/10.1177/0022034515572190
O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44(D1):D733–D745. https://doi.org/10.1093/nar/gkv1189
Brooks AJ, Waters MJ (2010) The growth hormone receptor: mechanism of activation and clinical implications. Nat Rev Endocrinol 6(9):515–525. https://doi.org/10.1038/nrendo.2010.123
Chen J, Chin A, Almarza AJ, Taboas JM (2020) Hydrogel to guide chondrogenesis versus osteogenesis of mesenchymal stem cells for fabrication of cartilaginous tissues. Biomed Mater 15(4):045006. https://doi.org/10.1088/1748-605X/ab401f
Davis MR, Summers KM (2012) Structure and function of the mammalian fibrillin gene family: implications for human connective tissue diseases. Mol Genet Metab 107(4):635–647. https://doi.org/10.1016/j.ymgme.2012.07.023
Li C, Cai Y, Chen S, Chen F (2016) Classification and characterization of class III malocclusion in Chinese individuals. Head Face Med 12(1):31. https://doi.org/10.1186/s13005-016-0127-8
Di Resta C, Ferrari M (2018) Next Generation Sequencing: From Research Area to Clinical Practice. EJIFCC 29(3):215–220
Jamuar SS, Tan EC (2015) Clinical application of next-generation sequencing for Mendelian diseases. Hum Genomics 9(1):10. https://doi.org/10.1186/s40246-015-0031-5
Hong EP, Park JW (2012) Sample size and statistical power calculation in genetic association studies. Genomics Inform 10(2):117–122. https://doi.org/10.5808/GI.2012.10.2.117
Xue F, Wong RW, Rabie AB (2010) Genes, genetics, and Class III malocclusion. Orthod Craniofacial Res 13(2):69–74. https://doi.org/10.1111/j.1601-6343.2010.01485.x
El-Gheriani AA, Maher BS, El-Gheriani AS, Sciote JJ, Abu-Shahba FA, Al-Azemi R, Marazita ML (2003) Segregation analysis of mandibular prognathism in Libya. J Dent Res 82(7):523–527. https://doi.org/10.1177/154405910308200707
Wolff G, Wienker TF, Sander H (1993) On the genetics of mandibular prognathism: analysis of large European noble families. J Med Genet 30(2):112–116. https://doi.org/10.1136/jmg.30.2.112
Nievergelt CM, Libiger O, Schork NJ (2007) Generalized analysis of molecular variance. PLoS Genet 3(4):e51. https://doi.org/10.1371/journal.pgen.0030051
Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, Bastarache LA, Wei WQ, Denny JC, Lin M, Hveem K, Kang HM, Abecasis GR, Willer CJ, Lee S (2018) Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet 50(9):1335–1341. https://doi.org/10.1038/s41588-018-0184-y
Author information
Authors and Affiliations
Contributions
Alexandra Dehesa-Santos contributed to the study conception and design, wrote the paper, and performed the literature search and data analysis of the manuscript.
Paula Iber-Diaz contributed to the literature search and data analysis.
Alejandro Iglesias-Linares contributed to the study conception and design and wrote the paper as well as its critical revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Online Resource 1.
Search strategy. (DOCX 15 kb)
Online Resource 2.
Full text excluded articles and reasons for exclusion. (DOCX 19 kb)
Online Resource 3.
Genetic variants contributing to Class III craniofacial characteristics. (PPTX 44 kb)
Online Resource 4.
Hardy-Weinberg equilibrium test for each genetic variant within eligible studies. (PPTX 700 kb)
Online Resource 5.
Egger's test Funnel Plots, sensitivity-Leave-1-out Forest plots random or fixed effects for each of the genetic variant. (PPTX 4440 kb)
Rights and permissions
About this article
Cite this article
Dehesa-Santos, A., Iber-Diaz, P. & Iglesias-Linares, A. Genetic factors contributing to skeletal class III malocclusion: a systematic review and meta-analysis. Clin Oral Invest 25, 1587–1612 (2021). https://doi.org/10.1007/s00784-020-03731-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-020-03731-5