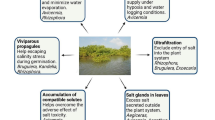
Abstract
Transcriptional regulatory networks are pivotal components of plant’s response to salt stress. However, plant adaptation strategies varied as a function of stress intensity, which is mainly modulated by climate change. Here, we determined the gene regulatory networks based on transcription factor (TF) TF_gene co-expression, using two transcriptomic data sets generated from the salt-tolerant “Tebaba” roots either treated with 50 mM NaCl (mild stress) or 150 mM NaCl (severe stress). The analysis of these regulatory networks identified specific TFs as key regulatory hubs as evidenced by their multiple interactions with different target genes related to stress response. Indeed, under mild stress, NAC and bHLH TFs were identified as central hubs regulating nitrogen storage process. Moreover, HSF TFs were revealed as a regulatory hub regulating various aspects of cellular metabolism including flavonoid biosynthesis, protein processing, phenylpropanoid metabolism, galactose metabolism, and heat shock proteins. These processes are essentially linked to short-term acclimatization under mild salt stress. This was further consolidated by the protein–protein interaction (PPI) network analysis showing structural and plant growth adjustment. Conversely, under severe salt stress, dramatic metabolic changes were observed leading to novel TF members including MYB family as regulatory hubs controlling isoflavonoid biosynthesis, oxidative stress response, abscisic acid signaling pathway, and proteolysis. The PPI network analysis also revealed deeper stress defense changes aiming to restore plant metabolic homeostasis when facing severe salt stress. Overall, both the gene co-expression and PPI network provided valuable insights on key transcription factor hubs that can be employed as candidates for future genetic crop engineering programs.




Similar content being viewed by others
Data availability
The RNA-seq datasets by using the Illumina-Hiseq platform are available from the NCBI Sequence Read Archive database (SRA; http://www.ncbi.nlm.nih.gov/sra) under project number accession PRJNA507974. The cDNA libraries were obtained from the controls and their respective salt-stress of 50 mM NaCl and 150 mM NaCl for 24 h with two biological replicates, respectively.
Abbreviations
- ABA:
-
Abscisic acid
- HSP:
-
Heat shock protein
- ER:
-
Endoplasmic reticulum
- TF:
-
Transcription factor
- ROS:
-
Reactive oxygen species
References
Ali A, Pardo JM, Yun D-J (2020) Desensitization of ABA-signaling: the swing from activation to degradation. Front Plant Sci 11:379
Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155(2):916–931
Arif Y, Singh P, Siddiqui H, Bajguz A, Hayat S (2020) Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Biochem 156:64–77
Askri H, Gharbi F, Rejeb S, Mliki A, Ghorbel A (2018) Differential physiological responses of Tunisian wild grapevines (Vitis vinifera L. subsp. sylvestris) to NaCl salt stress. Braz J Bot 41(4):795–804
Baena-González E (2010) Energy signaling in the regulation of gene expression during stress. Mol Plant 3(2):300–313
Balasubramaniam T, Shen G, Esmaeili N, Zhang H (2023) Plants&rsquo response mechanisms to salinity stress. Plants 12(12):2253
Baneh HD, Hassani A, Shaieste FG (2014) Effects of salinity on leaf mineral composition and salt injury symptoms of some Iranian wild grapevine (Vitis vinifera L. ssp. sylvestris) genotypes. OENO One 48(4):231–235
Bian S, Li R, Xia S, Liu Y, Jin D, Xie X, Dhaubhadel S, Zhai L, Wang J, Li X (2018) Soybean CCA1-like MYB transcription factor GmMYB133 modulates isoflavonoid biosynthesis. Biochem Biophys Res Commun 507(1–4):324–329
Bian XH, Li W, Niu CF, Wei W, Hu Y, Han JQ, Lu X, Tao JJ, Jin M, Qin H (2020) A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol 225(1):268–283
Budič M, Sabotič J, Meglič V, Kos J, Kidrič M (2013) Characterization of two novel subtilases from common bean (Phaseolus vulgaris L.) and their responses to drought. Plant Physiol Biochem 62:79–87
Cao H, Li J, Ye Y, Lin H, Hao Z, Ye N, Yue C (2020) Integrative transcriptomic and metabolic analyses provide insights into the role of trichomes in tea plant (Camellia sinensis). Biomolecules 10(2):311
Carillo P, Mastrolonardo G, Nacca F, Parisi D, Verlotta A, Fuggi A (2008) Nitrogen metabolism in durum wheat under salinity: accumulation of proline and glycine betaine. Funct Plant Biol 35(5):412–426
Carrasco D, Zhou-Tsang A, Rodriguez-Izquierdo A, Ocete R, Revilla MA, Arroyo-García R (2022) Coastal wild grapevine accession (Vitis vinifera L. ssp. sylvestris) shows distinct late and early transcriptome changes under salt stress in comparison to commercial rootstock Richter 110. Plants 11(20):2688
Chu S, Wang J, Zhu Y, Liu S, Zhou X, Zhang H, Wang C-e, Yang W, Tian Z, Cheng H (2017) An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet 13(5):e1006770
Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D (2014) What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol 165(2):519–527
Cochetel N, Ghan R, Toups HS, Degu A, Tillett RL, Schlauch KA, Cramer GR (2020) Drought tolerance of the grapevine, Vitis champinii cv. Ramsey, is associated with higher photosynthesis and greater transcriptomic responsiveness of abscisic acid biosynthesis and signaling. BMC Plant Biol. 20(1):1–25
Cramer GR, Van Sluyter SC, Hopper DW, Pascovici D, Keighley T, Haynes PA (2013) Proteomic analysis indicates massive changes in metabolism prior to the inhibition of growth and photosynthesis of grapevine (Vitis viniferaL.) in response to water deficit. BMC Plant Biol. 13(1):1–22
Cui MH, Yoo KS, Hyoung S, Nguyen HTK, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS (2013) An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett 587(12):1773–1778
Dalal M, Inupakutika M (2014) Transcriptional regulation of ABA core signaling component genes in sorghum (Sorghum bicolor L. Moench). Mol. Breed. 34(3):1517–1525
Daldoul S, Boubakri H, Gargouri M, Mliki A (2020) Recent advances in biotechnological studies on wild grapevines as valuable resistance sources for smart viticulture. Mol Biol Rep 47(4):3141–3153
Daldoul S, Hanzouli F, Hamdi Z, Chenenaoui S, Wetzel T, Nick P, Mliki A, Gargouri M (2022) The root transcriptome dynamics reveals new valuable insights in the salt-resilience mechanism of wild grapevine (Vitis vinifera subsp. sylvestris). Front Plant Sci 13:1077710
Daldoul S, Gargouri M, Weinert C, Jarrar A, Egert B, Mliki A, Nick P (2023) A Tunisian wild grape leads to metabolic fingerprints of salt tolerance. Plant Physiol 193(1):371–388. https://doi.org/10.1093/plphys/kiad304
Davila Olivas NH, Coolen S, Huang P, Severing E, van Verk MC, Hickman R, Wittenberg AH, de Vos M, Prins M, van Loon JJ (2016) Effect of prior drought and pathogen stress on Arabidopsis transcriptome changes to caterpillar herbivory. New Phytol 210(4):1344–1356
Diaz-Mendoza M, Velasco-Arroyo B, Santamaria ME, González-Melendi P, Martinez M, Diaz I (2016) Plant senescence and proteolysis: two processes with one destiny. Genet Mol Biol 39:329–338
Doncheva NT, Morris JH, Gorodkin J, Jensen LJ (2018) Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 18(2):623–632
Dong C, Zhang Z, Qin Y, Ren J, Huang J, Wang B, Lu H, Cai B, Tao J (2013) VaCBF1 from Vitis amurensis associated with cold acclimation and cold tolerance. Acta Physiol Plant 35(10):2975–2984
Fisarakis I, Chartzoulakis K, Stavrakas D (2001) Response of Sultana vines (V. vinifera L.) on six rootstocks to NaCl salinity exposure and recovery. Agric Water Manage 51(1):13–27
Foyer CH, Ferrario-Méry S, Noctor G (2001) Interactions between carbon and nitrogen metabolism. In: Lea PJ, Morot-Gaudry J-F (eds) Plant Nitrogen. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 237–254. https://doi.org/10.1007/978-3-662-04064-5_9
García-Calderón M, Pérez-Delgado CM, Palove-Balang P, Betti M, Márquez AJ (2020) Flavonoids and isoflavonoids biosynthesis in the model legume lotus japonicus; connections to nitrogen metabolism and photorespiration. Plants 9(6):774
Gargouri M, Park J-J, Holguin FO, Kim M-J, Wang H, Deshpande RR, Shachar-Hill Y, Hicks LM, Gang DR (2015) Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J Exp Bot 66(15):4551–4566
Grimplet J, Pimentel D, Agudelo-Romero P, Martinez-Zapater JM, Fortes AM (2017) The LATERAL ORGAN BOUNDARIES Domain gene family in grapevine: genome-wide characterization and expression analyses during developmental processes and stress responses. Sci Rep 7(1):15968
Ha J, Kang Y-G, Lee T, Kim M, Yoon MY, Lee E, Yang X, Kim D, Kim Y-J, Lee TR (2019) Comprehensive RNA sequencing and co-expression network analysis to complete the biosynthetic pathway of coumestrol, a phytoestrogen. Sci Rep 9 (1):1934
Han Z, Ahsan M, Adil MF, Chen X, Nazir MM, Shamsi IH, Zeng F, Zhang G (2020) Identification of the gene network modules highly associated with the synthesis of phenolics compounds in barley by transcriptome and metabolome analysis. Food Chem 323:126862
Hannah L, Roehrdanz PR, Ikegami M, Shepard AV, Shaw MR, Tabor G, Zhi L, Marquet PA, Hijmans RJ (2013) Climate change, wine, and conservation. Proc Natl Acad Sci 110(17):6907–6912
He X, Qu B, Li W, Zhao X, Teng W, Ma W, Ren Y, Li B, Li Z, Tong Y (2015) The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 169(3):1991–2005
Holsteens K, De Jaegere I, Wynants A, Prinsen EL, Van de Poel B (2022) Mild and severe salt stress responses are age-dependently regulated by abscisic acid in tomato. Front Plant Sci 13:982622
Hou H, Jia H, Yan Q, Wang X (2018) Overexpression of a SBP-box gene (VpSBP16) from Chinese wild Vitis species in Arabidopsis improves salinity and drought stress tolerance. Int J Mol Sci 19(4):940
Ivushkin K, Bartholomeus H, Bregt AK, Pulatov A, Kempen B, De Sousa L (2019) Global mapping of soil salinity change. Remote Sens Environ 231:111260
Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42(D1):D1182–D1187
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20(9):586–594
Jung C, Nguyen NH, Cheong J-J (2020) Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. Int J Mol Sci 21(24):9517
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32(suppl_1):D277–D280
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Ko DK, Brandizzi F (2020) Network-based approaches for understanding gene regulation and function in plants. Plant J 104(2):302–317
Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35(suppl_2):W345–W349
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147(1):316–330
Li H, Xu Y, Xiao Y, Zhu Z, Xie X, Zhao H, Wang Y (2010) Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232(6):1325–1337
Li X-W, Li J-W, Zhai Y, Zhao Y, Zhao X, Zhang H-J, Su L-T, Wang Y, Wang Q-Y (2013) A R2R3-MYB transcription factor, GmMYB12B2, affects the expression levels of flavonoid biosynthesis genes encoding key enzymes in transgenic Arabidopsis plants. Gene 532(1):72–79
Liao JC, Boscolo R, Yang Y-L, Tran LM, Sabatti C, Roychowdhury VP (2003) Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci 100(26):15522–15527
Liu X, Zhu Y, Zhai H, Cai H, Ji W, Luo X, Li J, Bai X (2012) AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem Biophys Res Commun 422(4):710–715
Liu Y, Wu Q, Qin Z, Huang J (2022) Transcription factor OsNAC055 regulates GA-mediated lignin biosynthesis in rice straw. Plant Sci 325:111455
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):1–21
Ma R, Xiao Y, Lv Z, Tan H, Chen R, Li Q, Chen J, Wang Y, Yin J, Zhang L (2017) AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Front Plant Sci 8:1361
Martinoia E, Rentsch D (1994) Malate compartmentation-responses to a complex metabolism. Annu Rev Plant Biol 45(1):447–467
Meiri D, Tazat K, Cohen-Peer R, Farchi-Pisanty O, Aviezer-Hagai K, Avni A, Breiman A (2010) Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol Biol 72:191–203
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nonami H, Wu Y, Boyer JS (1997) Decreased growth-induced water potential (a primary cause of growth inhibition at low water potentials). Plant Physiol 114(2):501–509
Pascual-Ahuir A, González-Cantó E, Juyoux P, Pable J, Poveda-Huertes D, Saiz-Balbastre S, Squeo S, Ureña-Marco A, Vanacloig-Pedros E, Zaragoza-Infante L (2019) Dose dependent gene expression is dynamically modulated by the history, physiology and age of yeast cells. Biochim Biophys Acta (BBA)-Gene Regul Mech 1862 (4):457–471
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295
Pillet J, Egert A, Pieri P, Lecourieux F, Kappel C, Charon J, Gomès E, Keller F, Delrot S, Lecourieux D (2012) VvGOLS1 and VvHsfA2 are involved in the heat stress responses in grapevine berries. Plant Cell Physiol 53(10):1776–1792
Rao X, Dixon RA (2019) Co-expression networks for plant biology: why and how. Acta Biochim Biophys Sin 51(10):981–988
Raymond P, Brouquisse R, Chevalier C, Couée I, Dieuaide M, James F, Just D, Pradet A (1994) Proteolysis and proteolytic activities in the acclimation to stress: the case of sugar starvation in maize root tips. In: Biochemical and Cellular Mechanisms of Stress Tolerance in Plants. Berlin, Heidelberg, pp 325–334
Ren C, Kuang Y, Lin Y, Guo Y, Li H, Fan P, Li S, Liang Z (2022) Overexpression of grape ABA receptor gene VaPYL4 enhances tolerance to multiple abiotic stresses in Arabidopsis. BMC Plant Biol 22(1):1–14
Sarath NG, Sruthi P, Shackira AM, Puthur JT (2021) Chapter 16 - Halophytes as effective tool for phytodesalination and land reclamation. In: Aftab T, Hakeem KR (eds) Frontiers in Plant-Soil Interaction. Academic Press, pp 459–494. https://doi.org/10.1016/B978-0-323-90943-3.00020-1
Schaller A, Stintzi A, Graff L (2012) Subtilases–versatile tools for protein turnover, plant development, and interactions with the environment. Physiol Plant 145(1):52–66
Scheible W-R, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9(5):783–798
Serrano I, Campos L, Rivas S (2018) Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front Plant Sci 9:139
Shahmoradi M, Shekafandeh A, Eshghi S (2019) Physiological and biochemical changes of some grapevine cultivars under different irrigation regimes. Agric Conspec Sci 84(4):371–382
Shavrukov Y (2013) Salt stress or salt shock: which genes are we studying? J Exp Bot 64(1):119–127
Silveira J, Melo A, Viégas R, Oliveira J (2001) Salinity-induced effects on nitrogen assimilation related to growth in cowpea plants. Environ Exp Bot 46(2):171–179
Singh M, Kumar J, Singh V, Prasad S (2014) Plant tolerance mechanism against salt stress: the nutrient management approach. Biochem. Pharmacol 3(165):2167–0501.1000
Singh M, Singh VP, Prasad SM (2019) Nitrogen alleviates salinity toxicity in Solanum lycopersicum seedlings by regulating ROS homeostasis. Plant Physiol Biochem 141:466–476
Song C-P, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu J-K (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17(8):2384–2396
Stitt M, Scheible W-R (1999) Nitrate acts as a signal to control gene expression, metabolism and biomass allocation. In: Kruger NJ, Hill SA, Ratcliffe RG (eds) Regulation of Primary Metabolic Pathways in Plants. Springer Netherlands, Dordrecht, pp 275–306. https://doi.org/10.1007/978-94-011-4818-4_14
Tai Y, Liu C, Yu S, Yang H, Sun J, Guo C, Huang B, Liu Z, Yuan Y, Xia E (2018) Gene co-expression network analysis reveals coordinated regulation of three characteristic secondary biosynthetic pathways in tea plant (Camellia sinensis). BMC Genom 19:1–13
Tani E, Sarri E, Goufa M, Asimakopoulou G, Psychogiou M, Bingham E, Skaracis GN, Abraham EM (2018) Seedling growth and transcriptional responses to salt shock and stress in Medicago sativa L., Medicago arborea L., and Their Hybrid (Alborea). Agronomy 8(10):231
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9(5):244–252
Wong DC, Matus JT (2017) Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: recent applications and future opportunities. Front Plant Sci 8:505
Wu H, Hill CB, Stefano G, Bose J (2021a) New insights into salinity sensing, signaling and adaptation in plants, vol 11. Frontiers Media SA
Wu H, Hill CB, Stefano G, Bose J (2021b) Editorial: New insights into salinity sensing, signaling and adaptation in plants. Front Plant Sci 11. https://doi.org/10.3389/fpls.2020.604139
Xu FQ, Xue HW (2019) The ubiquitin-proteasome system in plant responses to environments. Plant, Cell Environ 42(10):2931–2944
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J Integrative Biol 16(5):284–287
Yuan Y, Qin L, Su H, Yang S, Wei X, Wang Z, Zhao Y, Li L, Liu H, Tian B (2021) Transcriptome and coexpression network analyses reveal hub genes in Chinese cabbage (Brassica rapa L. ssp. pekinensis) during different stages of Plasmodiophora brassicae infection. Frontiers in Plant Science 12:650252
Zhang H, Zhao Y, Zhu J-K (2020) Thriving under stress: how plants balance growth and the stress response. Dev Cell 55(5):529–543
Zhao D, Derkx A, Liu DC, Buchner P, Hawkesford M (2015) Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol 17(4):904–913
Zhu Z, Shi J, Cao J, He M, Wang Y (2012) VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep 31(11):2109–2120
Zhu Z, Shi J, Xu W, Li H, He M, Xu Y, Xu T, Yang Y, Cao J, Wang Y (2013) Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J Plant Physiol 170(10):923–933
Zou L, Zhong G-Y, Wu B, Yang Y, Li S, Liang Z (2019) Effects of sunlight on anthocyanin accumulation and associated co-expression gene networks in developing grape berries. Environ Exp Bot 166:103811
Funding
This research was carried out as part of the Tunisian National Research Program (CP2019-2022) of the Laboratory of Plant Molecular Physiology at the Centre of Biotechnology of Borj-Cedria, funded by the Tunisian Ministry of Higher Education and Scientific Research.
Author information
Authors and Affiliations
Contributions
S.D. and M.G. conceived and designed the study. F.H. and S.D performed the physiological analyses. S.D. performed the molecular analyses. M.G. performed the bioinformatic and statistical analyses. H.B. performed PPI interaction networks. S.D. and M.G wrote the original draft of the manuscript. All authors read and contribute to the review and editing of the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Handling Editor: Reimer Stick
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Daldoul, S., Hanzouli, F., Boubakri, H. et al. Deciphering the regulatory networks involved in mild and severe salt stress responses in the roots of wild grapevine Vitis vinifera spp. sylvestris. Protoplasma 261, 447–462 (2024). https://doi.org/10.1007/s00709-023-01908-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-023-01908-9