Abstract
Glucose deprivation in the slime mold Physarum polycephalum leads to a specific morphotype, a highly motile mesoplasmodium. We investigated the ultrastructure of both mesoplasmodia and non-starved plasmodia and found significantly increased numbers of mitochondria in glucose-deprived mesoplasmodia. The volume of individual mitochondria was the same in both growth forms. We conjecture that the number of mitochondria correlates with the metabolic state of the cell: When glucose is absent, the slime mold is forced to switch to different metabolic pathways, which occur inside mitochondria. Furthermore, a catabolic cue (such as AMP-activated protein kinase (AMPK)) could stimulate mitochondrial biogenesis.
Similar content being viewed by others
Introduction
The giant unicellular slime mold Physarum polycephalum grows into transport networks (termed macroplasmodia) which can reach sizes of up to square meters (Stockem and Brix 1994). Under certain nutritional conditions, i.e., a lack of glucose in the solid agar medium, and when the culture has reached a certain age, a special foraging pattern can be observed. Instead of forming a coherent network from isolated fragments, the slime mold aggregates into independent, unconnected units (termed mesoplasmodia), which then move in a straight trajectory away from their point of origin (Lee et al. 2018). Mesoplasmodia are well suited as models to study the mechanism of the slime mold’s locomotion (Oettmeier and Döbereiner 2019), because they move for hours in straight trajectories and keep a constant shape. The movement of those autonomous foraging units is comparatively fast, reaching speeds of up to 17 μm/min.
Slime molds exhibit characteristic continuous rhythmic oscillations, orchestrated by the cytoskeletal proteins actin and myosin. A detailed description of the ultrastructure can be found in Oettmeier et al. (2018). These vigorous and perpetual contractions, which are the basis for locomotion in P. polycephalum (Oettmeier and Döbereiner 2019), require a lot of energy in the form of ATP, which is supplied by glycolysis and oxidative phosphorylation in mitochondria. Glycolysis takes place in the cytoplasm and seems to be intensely operated by the slime mold (Sauer 1982).
P. polycephalum possesses mitochondria with tubular cristae. The ultrastructure of eumycetozoan mitochondria is unique and characteristic for slime molds (Dykstra 1977), but their function is the same as in any other eukaryotic organism. Mitochondria in P. polycephalum are isolated and spherical or lenticular (see Fig. 1).
Our observations of the shape conform to earlier findings (Daniel and Järlfors 1972; Sauer 1982). Mitochondria do not form networks, because the vigorous intracellular flow within the amoeboid cell body is constantly moving them around. A video of this can be found in the supplementary material (Video S1a and S1b). An elaborate mitochondrial network (as for example in budding yeast) would not be feasible due to the dynamic nature of the cytoplasm. Furthermore, it has been demonstrated that the slime mold’s mitochondria can migrate within the cell (Kuroiwa and Takahashi 1978) in response to culture conditions: they moved towards the periphery of the cell when a liquid shaking culture was left unstirred for a few hours. This migration is reversible; when the microplasmodia were agitated, they dispersed evenly again. Apart from the characteristic tubular cristae, the mitochondria possess mitochondrial DNA (mtDNA), which is packaged into the electron-dense mitochondrial nucleoid (see Fig. 1a), along with many proteins (Itoh et al. 2011). The complete mitochondrial genome has been sequenced (Takano et al. 2001).
Mitochondria perform many important biological functions. Most important is the production of ATP through oxidative phosphorylation, but they also play a role in the pronounced and well-described oscillations of the slime mold: mitochondria store and release calcium (Nations et al. 1989; Achenbach et al. 1984), thereby forming a crucial component of the biochemical oscillator. The nature and localization of this pacemaker of the contraction-relaxation cycle poses one of the most interesting problems regarding the dynamic processes of the non-muscle contractile system in P. polycephalum. Although the exact mechanism remains unknown, it is clear that mitochondria and the processes taking place within them are integral parts (Satoh et al. 1982; Korohoda et al. 1983). Inhibiting glycolysis or respiration leads to changes in the pattern and frequency of the oscillations.
When sufficient glucose is present in the medium, glycolysis takes place in the cytosol, producing two molecules of ATP and two molecules of NADH per molecule of glucose. Furthermore, it produces two molecules of pyruvate which are then transported into the mitochondria to enter the citric acid cycle. Electrons from the glycolysis and citric acid cycle are then being transferred by NADH and FADH2 to the electron transport chain, ultimately driving oxidative phosphorylation and producing more ATP (more than 30 molecules per molecule of glucose). Citric acid cycle and oxidative phosphorylation take place across the inner membrane and cristae of the mitochondria. When glucose is absent from the medium, P. polycephalum immediately starts to use its abundant stores of glycogen in a process termed glycogenolysis (Nader and Becker 1983). The glycogen polymer is broken down by the enzyme glycogen phosphorylase, releasing glucose, which can then be used in glycolysis. Besides glucose (and other carbohydrates), the slime mold is also able to catabolize proteins (Goodman and Beck 1974) and lipids (Poulos and Thompson 1971). If given a choice, P. polycephalum seems to prefer a diet that contains equal ratios of proteins and carbohydrates, or a ratio of two times more proteins than carbohydrates (Dussutour et al. 2010).
Usually, when microplasmodia of P. polycephalum are cultivated in a growth medium that lacks nutrients (but contains salts to maintain pH), inactive cyst-like stages are formed after a certain time (Hüttermann 1973). These spherules also occur when a shaking culture of microplasmodia depletes its liquid medium of nutrients. Depending on the starting conditions (temperature, culture volume, nutrient concentrations, shaking speed), glucose is depleted after 2.5 (Nader and Becker 1983) to 4 days (Lee et al. 2018). In this study, we used the same conditions as described in Lee et al. (2018), which means that microplasmodia reach their maximum biomass after 3 to 4 days and turn into spherules after ~ 7 days after inoculation, if left in their shaking culture. However, when microplasmodia from a 6-day-old culture are plated onto an agar plate lacking glucose, they will form the aforementioned mesoplasmodia which then begin to move outward from the inoculation center. For about 9 h, the mesoplasmodia migrate on straight trajectories without much changing their shape or showing an increase in biomass. Our samples were taken from mesoplasmodia in the middle of the migration period. After this motile phase, at around 10 h after initial plating, the mesoplasmodia reach a pause state in which migration is ceased. After this pause, mesoplasmodia either transition into static networks, continue to migrate, or move in a different pattern.
As observed in skeletal muscle cells, an elevated energy demand (e.g., through exercise) increases mitochondrial volume density (Lundby and Jacobs 2016). Similarly, myocardial hypertrophy (Wiesner et al. 1994) changes in neuronal activity (Liu and Wong-Riley 1995), and other metabolic challenges lead to an increase in mitochondrial biogenesis. Therefore, many cells are capable of adjusting their mitochondria to a change of energy demand, requiring that they possess an appropriate intracellular energy sensor. Since mitochondria are so important for both energy metabolism and the primary oscillator, we compared, in the present study, the mitochondria of glucose-deprived mesoplasmodia and non-starved plasmodia. We found a significantly increased number of mitochondria in glucose-deprived mesoplasmodia. We hypothesize that mitochondrial biogenesis is stimulated in P. polycephalum mesoplasmodia grown in the absence of glucose, probably in order to compensate for the reduced supply of glycolytic ATP and pyruvate.
Material and methods
Microplasmodia culture
We used the strain WT33 (Marwan and Starostzik 2002) × LU898 (Kawano et al. 1987), which was kindly provided by Prof. Dr. Wolfgang Marwan (Universität Magdeburg). Microplasmodia were grown in a liquid growth medium (see Tables 1 and 2). The cultures were grown at a constant temperature of 24 °C and rotation speed (180 rpm) in the dark. Torn apart by shear forces, multiple small and spherical units are produced, whose size is determined by the shaking speed. Fresh microplasmodia cultures were prepared by taking 2 ml of the previous culture at days 3 to 4, centrifuging gently and discarding the supernatant. The pellet was then transferred into new liquid medium.
Mesoplasmodia
To create mesoplasmodia, microplasmodia from a 6-day-old liquid culture were centrifuged, the supernatant discarded, and they were resuspended briefly with MilliQ water. Microplasmodia were then transferred onto a semi-defined medium (SDM) agar plate lacking glucose (see Table 3).
P. polycephalum requires hemin to grow (Daniel et al. 1962). However, hemin is poorly soluble in water. Therefore, it needs to be dissolved in 1 N NaOH first. This solution can then be mixed with MilliQ water to achieve the desired final concentration.
SDM agar contains an additional 20 g of D(+) glucose per liter when used to grow typical macroplasmodial networks. On the glucose-deficient agar, the microplasmodia form aggregates and fuse with each other. After about 3 h, the first migrating units leave the initial patch and move radially outwards.
Transmission electron microscopy
Macroplasmodia and mesoplasmodia growing on an agar surface were submerged with a fixative (80 mM KCl, 50 mM sodium cacodylate, pH 7.2, 20 mM NaCl, 2.5% glutaraldehyde). The fixation was carried out for at least 30 min on ice. Plasmodia were fixed together with the agar they were growing on, which was then cut into blocks the size of a few square millimeters with a scalpel. Post-fixation and contrast enhancement was carried out in 2% OsO4 on ice for 60 min. After post-fixation, the samples were washed thoroughly with double-distilled water. Samples were then left to contrast for 12 h at 4 °C in the dark in 0.5% uranyl acetate. Uranyl acetate helps to increase the contrast as well as the stability of the fine structures of the cell. After fixing and contrasting, the samples were dehydrated by passing them through a series of increasing ethanol concentrations. First, they were treated with 30% ethanol for 30 min, whereby the ethanol was exchanged after 15 min. This procedure was repeated for 50%, 70%, 90%, and 100% ethanol. An extra step was performed with 100% ethanol, dehydrated with a molecular sieve (15 min).
Next, the specimens were embedded in resin. Embedding was performed with glycid ether 100 (Luft 1961). The ethanol was replaced in a descending alcohol series with the glycid ether 100. The glycid ether solutions A and B were mixed at a ratio 3:7 (A:B). A descending alcohol series was prepared as follows at room temperature: First, the samples were infiltrated for 15 min with a mixture of 100% ethanol and glycid ether A + B at a ratio of 3:1, then at a 1:1 ratio for 15 min, and finally at a 1:3 ratio for 15 min. The ethanol-resin mixture was carefully removed and the last step was infiltration of the tissue with a pure glycid ether (A + B) mixture for a total of 45 min, whereby the glycid ether was replaced twice after 15 min each. After the glycid ether was removed for the last time, the accelerator DMP-30 was added to the mixture. The samples were then left to polymerize in a vacuum oven at 60 °C for 2 to 3 days. Ultra-thin cutting (40–60 nm) was performed with a Reichert-Jung Ultracut E microtome. TEM was carried out on a Zeiss EM 900, equipped with a water-cooled frame-transfer-CCD-camera (TRS). Images were acquired using a PC with the software “ImageSP” (TRS).
Stereological measurements
Stereology is defined as a set of mathematical methods which relate parameters defining three-dimensional structures to measurements obtained from two-dimensional sections. In other words, one can estimate higher dimensional information from lower dimensional samples. The advantage of evaluating thin sections with stereology is that it yields quantitative morphological data. Geometric properties of structures (e.g., mitochondria) embedded in a referent space (e.g., cytoplasm) can be estimated by studying the intersection of these structures with a probe. Probes are points, lines, or grids which are being superimposed onto the section (image). A prerequisite for stereology is that samples must be uniform, isotropic, and random (UIR); this means that the orientation of the cut surface and the position of the embedded specimen must be random, as well as the positioning of probes. We use the following stereological relationships:
Volume density
Volume density (VV) or volume fraction is the ratio between the volume of the structure and the volume of the referent space (Eq. 1).
The probes used are points. A point grid is superimposed onto the TEM image and mitochondria are counted which coincide with the probes (see Fig. 2a).
a Random offset grid. The number of points (red crosses) which fall on mitochondria is counted (Pi), as well as the number of points which falls onto the referent volume (cytoplasm, Qi). This takes into account the relatively high porosity of the slime mold’s cytoplasm. Scale bar = 2 μm. b Counting frame. Green line = inclusion line, red line = exclusion line. Scale bar = 2 μm
By doing a point count on an image (i), we obtain the number of points which fall onto mitochondria (Pi) and the number of points in the referent space (i.e., cytoplasm, Qi). The volume density states which percentage of the cytoplasm is occupied by mitochondria. FIJI provides grids with random offset. Depending on magnification, the area per point was chosen to lie between 1 and 5 μm2 (110–361 points per image).
Numerical density
Numerical density (NV) is the number of structures per volume of the referent space (i.e., number of mitochondria per unit volume of cytoplasm). In this case, the probes are volumes. Since the mitochondria of P. polycephalum are ellipsoid in shape and of similar sizes, we can use the method proposed by Weibel and Gomez (1962). First, using a counting frame (a macro (Mironov 2014) implemented in FIJI), the number of structures per unit area (NA) is computed (see Fig. 2b). Following stereological rules, a mitochondrion is only counted if it lies entirely within the counting frame or if it touches a green inclusion line. It is not counted if it intersects a red exclusion line. Second, we need to calculate ϵ, i.e., the ratio of short (a) to long semi-axis (b) (Eq. 2).
Here, ϵ is approximately 0.86, indicating a slightly prolate ellipsoid (ϵ < 1). For each ϵ, the corresponding shape factor β has to be obtained from literature (Weibel and Gomez 1962). In our case, β is 1.4. For a perfect sphere, ϵ = 1 and β = 1.38. The numerical density (NV) can now be calculated using Eq. 3.
Mean mitochondrial volume
The mean volume \( \overline{V} \) of mitochondria can be calculated from VV and NV using Eq. 4 (Cruz-Orive and Weibel 1990):
Autofluorescence
Autofluorescence imaging of microplasmodia was performed using a Zeiss Axio Oberver.Z1 equipped with a Zeiss incubation system consisting of Heating Unit XL S and Temp Module S. Imaging was carried out at 24 °C. A Zeiss Plan Apochromat × 40 with a numerical aperture of 0.95 was used, and images were taken by a Zeiss Axio-Cam MRm. Microplasmodia were plated onto Petri dishes with thin glass bottoms. They were illuminated at a wavelength of 380 nm with a Zeiss HXP 120 mercury lamp, of which the UV filter was removed. We used a 79000 ET FURA 2 Hybrid filter set (Chroma) and a Zeiss 76 HE reflector filter set.
Results
Volume fraction, number density, and mean volume
We compared TEM images of three specimens each of glucose-deprived mesoplasmodia and non-starved plasmodia. On average, mitochondria in unstarved plasmodia occupy ~ 4% of the cytoplasm volume, but ~ 9% in starved mesoplasmodia (Fig. 3).
a Volume fraction (VV) of mitochondria in unstarved (blue) and starved (red) slime mold. Starved mesoplasmodia contain a significantly higher volume fraction of mitochondria (p < 0.001) than unstarved specimens. b Numerical density (NV) [μm−3] of mitochondria in unstarved and starved slime mold. Again, the difference is statistically significant (p < 0.001). Error bars = standard deviation σ
The results for the numerical density (VV) are similar: Unstarved plasmodia contain ~ 0.035 mitochondria per μm−3, whereas starved mesoplasmodia contain ~ 0.08 mitochondria per μm−3. There are approximately twice as many mitochondria per unit volume of cytoplasm in mesoplasmodia than in unstarved plasmodia. Both the results for VV and NV show statistically highly significant differences as confirmed by two-sample T tests (both p < 0.001). The mean mitochondrial volume, however, does not vary between starved and unstarved plasmodia (see Fig. 4).
Autofluorescence
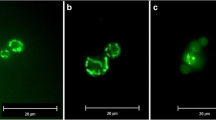
When living plasmodia are illuminated with wavelengths in the range of 340 to 380 nm, they show pronounced autofluorescence with an emission wavelength of around 460 nm. A base autofluorescence is detectable in the cytoplasm, as well as brightly fluorescing spots (see Fig. 5b).
We deduce that these spots are mitochondria. This conclusion is based on the presence of NAD and its reduced form, NADH, in both mitochondria and cytoplasm. NADH strongly absorbs ultraviolet light, with an emission peak at 460 nm. In small amounts, NADH is produced during glycolysis, which explains the low fluorescence of the cytoplasm. However, the largest share of the cell’s NADH is found inside the mitochondria (Ince et al. 1992), accounting for the strong autofluorescence. The autofluorescence data also confirms our stereological finding that mitochondria do not form networks, but are rather isolated organelles (see Video S1b). NADH autofluorescence can be used to assess intracellular pH (Ogikubo et al. 2011), monitor mitochondrial toxicity (Rodrigues et al. 2011), and can generally give insight into the energy metabolism (Bartolomé and Abramov 2015; Evans et al. 2005; Mayevsky and Rogatsky 2007). However, in P. polycephalum, this autofluorescence can cause problems when short-wavelength calcium-staining dyes, such as Fura 2, are used. A video of a living microplasmodium exhibiting autofluorescence (Video S1b) and the same microplasmodium at bright field illumination (Video S1a) can be found in the supplementary material.
Discussion
Our results show that under glucose-deprived conditions, the number of mitochondria is significantly increased. Their volume does not differ between glucose-deprived and unstarved plasmodia, indicating that this is not an instance of fragmentation. In contrast, in mouse embryonic fibroblasts, glucose depletion leads to increased mitochondrial fragmentation (Rambold et al. 2011). Likewise, in yeast, glucose deprivation under aerobic conditions leads to a fragmentation of mitochondria into many small units (Visser et al. 1995). However, how cells respond in detail to glucose withdrawal is not well studied, and results are controversial (Song and Hwang 2019; Wappler et al. 2013). In cancer cells, for example, glucose deprivation causes cell death (Iurlaro et al. 2017). However, the ability to reprogram the energy metabolism is a hallmark of cancer (Hanahan and Weinberg 2011). In other cell types, viability is not significantly affected (Jelluma et al. 2006). It seems that there is a great variability in the response to glucose depletion, depending also on cofactors like a simultaneous lack of oxygen. For example, after a non-lethal phase of both oxygen and glucose depletions, an increase in mitochondrial biogenesis in neurons was observed (Wappler et al. 2013).
Apart from fragmentation, mitochondrial morphology can be affected by the metabolic state of the cell. Starved amoeba of the species Chaos carolinense exhibited highly organized special membrane structures within their mitochondria (Chong et al. 2018). During starvation-induced autophagy, mitochondria can increase in size and become elongated in shape, which optimizes ATP production and spares them from being digested (Blackstone and Chang 2011). In other words, stress can affect mitochondrial morphology. Our results show no difference in morphology between glucose-deprived and non-starved plasmodia. A difference to the studies cited above, however, is that apart from a lack of glucose, the medium contained a source of protein (see Table 3). Soytone is an enzymatic digest of soybean meal, and it contains peptides, amino acids, vitamins, and complex carbohydrates. Those are alternative energy sources that the slime mold can metabolize, and therefore, neither the increase in number nor the morphology is related to stress.
Our results show that mitochondrial biogenesis is stimulated in P. polycephalum grown in the absence of glucose, probably in order to compensate for the diminished supply of glycolytic ATP and pyruvate. We speculate that the number of mitochondria correlates to the metabolic state of the cell (see Fig. 6).
The increase in mitochondrial numbers leads to a higher ATP production. In conditions where glucose is abundant (upper panel in Fig. 6), glucose is converted to ATP and pyruvate via glycolysis. At the same time, the slime mold stores surplus energy in the form of glycogen (Goodman and Rusch 1969; Nader and Becker 1983). Pyruvate is then transported into the mitochondria, where it enters the citric acid cycle, and during oxidative phosphorylation, more ATP is produced. This seems to be the preferred metabolic pathway when sufficient glucose is present. However, when glucose is withdrawn (lower panel in Fig. 6), different metabolic pathways are taken. First, P. polycephalum uses up its glycogen storages. Glycogenolysis releases glucose from glycogen, which then enters glycolysis. Nader and Becker (1983) have measured that after the glucose in the growth medium was depleted, glycogen stores within microplasmodia lasted for a period of ~ 5.5 days until it ran out. As soon as exogenous glucose is consumed or removed, glycogen is degraded.
Another pathway during glucose depletion starts with lower levels of ATP in the cell, with a simultaneous increase in AMP. This is a metabolic cue, which leads to the activation of AMP-activated protein kinase (AMPK). This enzyme belongs to a highly conserved protein family with orthologs in yeast (Saccharomyces cerevisiae (SNF1)) (Hedbacker and Carlson 2008), in other fungi, in plants (SnRK1) (Margalha et al. 2016), and in Dictyostelium discoideum (Bokko et al. 2007), a member of the amoebozoa group of organisms to which Physarum also belongs. Several AMPK orthologs are encoded in the P. polycephalum genome (Schaap et al. 2016) and are expressed in starving, sporulation-competent plasmodia (Glöckner and Marwan 2017). AMPK plays a role in cellular energy homeostasis. When ATP levels lower, AMPK activation stimulates, among other processes, fatty acid oxidation and mitochondrial biogenesis (Mihaylova and Shaw 2011; Song and Hwang 2019). Furthermore, AMPK enhances protein catabolism (He et al. 2017). AMPK is an intracellular energy status sensor and key regulator of mitochondrial biogenesis. When activated by low ATP levels, AMPK triggers a metabolic switch, decreasing the activity of anabolic pathways and enhancing catabolic processes to restore the energy balance.
In summary, we propose that an imbalance between energy requirement and energy supply (deprivation of glucose) regulates mitochondrial biogenesis. By withdrawing glucose from the culture medium, we forced the slime mold to be exclusively dependent on mitochondrial ATP production. As a result, mitochondrial biogenesis was increased and we found a very high number of mitochondria. Additionally, mesoplasmodia are migrating fast and far in search for food, and this locomotion is also very energy consuming. To compensate for a lack of glycolytic ATP, mitochondrial numbers are increased. Our findings highlight the importance of the AMPK-like metabolic switch in P. polycephalum. This pathway has not yet been confirmed for glucose-deprived mesoplasmodia, but appears to be a very likely candidate to explain our observations. A closer investigation of this sophisticated system of energy metabolism adaptation is needed in order to get a more complete understanding of how P. polycephalum manages homeostasis in the face of nutritional challenges.
References
Achenbach F, Achenbach U, Kessler D (1984) Calcium binding sites in plasmodia of Physarum polycephalum as revealed by the pyroantimonate technique. J Histochem Cytochem 32(11):1177–1184
Bartolomé F, Abramov AY (2015) Measurement of mitochondrial NADH and FAD autofluorescence in live cells. Methods Mol Biol 1264:263–270
Blackstone C, Chang CR (2011) Mitochondria unite to survive. Nat Cell Biol 13(5):521–522
Bokko PB, Francione L, Bandala-Sanchez E, Ahmed AU, Annesley SJ, Huang X et al (2007) Diverse cytopathologies in mitochondrial disease are caused by AMP-activated protein kinase signaling. Mol Biol Cell 18(5):1874–1886
Chong K, Almsherqi ZA, Shen HM, Deng Y (2018) Cubic membrane formation supports cell survival of amoeba Chaos under starvation-induced stress. Protoplasma 255(2):517–525
Cruz-Orive LM, Weibel ER (1990) Recent stereological methods for cell biology: a brief survey. Am J Phys 258(4 Pt 1):L148–L156
Daniel JW, Järlfors U (1972) Plasmodial ultrastructure of the myxomycete Physarum polycephalum. Tissue Cell 4(1):15–36
Daniel JW, Kelley J, Rusch HP (1962) Hematin-requiring plasmodial myxomycete. J Bacteriol 84:1104–1110
Dussutour A, Latty T, Beekman M, Simpson SJ (2010) Amoeboid organism solves complex nutritional challenges. PNAS 107(10):4607–4611
Dykstra MJ (1977) The possible phylogenetic significance of mitochondrial configurations in the acrasid cellular slime molds with reference to members of the eumycetozoa and fungi. Mycologia 69(3):579–591
Evans ND, Gnudi L, Rolinski OJ, Birch DJ, Pickup JC (2005) Glucose-dependent changes in NAD(P)H-related fluorescence lifetime of adipocytes and fibroblasts in vitro: Potential for noninvasive glucose sensing in diabetes mellitus. J Photochem Photobiol B 80(2):122–129
Glöckner G, Marwan W (2017) Transcriptome reprogramming during developmental switching in Physarum polycephalum involves extensive remodeling of intracellular signaling networks. Sci Rep 7:12304
Goodman EM, Beck T (1974) Metabolism during differentiation in the slime mold Physarum polycephalum. Can J Microbiol 20(2):107–111
Goodman EM, Rusch HP (1969) Glycogen in Physarum polycephalum. Cell Mol Life Sci 25(6):580–580
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
He L, Zhou X, Huang N, Li H, Tian J, Li T, Yao K, Nyachoti CM, Kim SW, Yin Y (2017) AMPK regulation of glucose, lipid and protein metabolism: mechanisms and nutritional significance. Curr Protein Pept Sci 18(6):562–570
Hedbacker K, Carlson M (2008) SNF1/AMPK pathways in yeast. Front Biosci 13:2408
Hüttermann A (1973) Biochemical events during spherule formation of Physarum polycephalum. Ber Dtsch Botanischen Ges 86:55–76
Ince C, Coremans JM, Bruining HA (1992) In vivo NADH fluorescence. Adv Exp Med Biol 317:277–296
Itoh K, Izumi A, Mori T, Dohmae N, Yui R, Maeda-Sano K, Shirai Y, Kanaoka MM, Kuroiwa T, Higashiyama T, Sugita M, Murakami-Murofushi K, Kawano S, Sasaki N (2011) DNA packaging proteins Glom and Glom2 coordinately organize the mitochondrial nucleoid of Physarum polycephalum. Mitochondrion 11(4):575–586
Iurlaro R, Püschel F, Léon-Annicchiarico CL, O’Connor H, Martin SJ, Palou-Gramón D, Lucendo E, Muñoz-Pinedo C (2017) Glucose deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol Cell Biol 37(10):e00479–e00416
Jelluma N, Yang X, Stokoe D, Evan GI, Dansen TB, Haas-Kogan DA (2006) Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol Cancer Res 4:319–330
Kawano S, Andersen RW, Nanba T, Kuroiwa T (1987) Polymorphism and uniparental inheritance of mitochondrial DNA in Physarum polycephalum. J Gen Microbiol 133:3175–3182
Korohoda W, Shraideh Z, Baranowski Z, Wohlfarth-Bottermann KE (1983) Energy metabolic regulation of oscillatory contraction activity in Physarum polycephalum. Cell Tissue Res 231:675–669
Kuroiwa T, Takahashi K (1978) Induction of mitochondrial migration in the slime mold Physarum polycephalum. Plant Cell Physiol 19(8):1561–1564
Lee J, Oettmeier C, Döbereiner H-G (2018) A novel growth mode of Physarum polycephalum during starvation. J Phys D Appl Phys 51(24):244002
Liu S, Wong-Riley M (1995) Disproportionate regulation of nuclear- and mitochondrial-encoded cytochrome oxidase subunit proteins by functional activity in neurons. Neuroscience 67:197–210
Luft JH (1961) Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol 9(2):409–414
Lundby C, Jacobs RA (2016) Adaptations of skeletal muscle mitochondria to exercise training. Exp Physiol 101(1):17–22
Margalha L, Valerio C, Baena-González E (2016) Plant SnRK1 kinases: structure, regulation, and function. In: AMP-activated protein kinase, vol 107. Springer, Cham, pp 403–438
Marwan W, Starostzik C (2002) The sequence of regulatory events in the sporulation control network of Physarum polycephalum analysed by time-resolved somatic complementation of mutants. Protist 153:39–400
Mayevsky A, Rogatsky GG (2007) Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Phys Cell Physiol 292(2):C615–C640
Mihaylova MM, Shaw RJ (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13(9):1016–1023
Mironov A (2014) Unbiased frames. Version 1.0 https://imagej.nih.gov/ij/macros/Unbiased_Frames.txt
Nader WF, Becker JU (1983) Regulation of glycogen metabolism and glycogen phosphorylase in Physarum polycephalum. Microbiology 129(8):2481–2487
Nations C, Allison VF, Aldrich HC, Allen RG (1989) Biological oxidation and the mobilization of mitochondrial calcium during the differentiation of Physarum polycephalum. J Cell Physiol 140:311, 316
Oettmeier C, Döbereiner H-G (2019) A lumped parameter model of endoplasm flow in Physarum polycephalum explains migration and polarization-induced asymmetry during the onset of locomotion. PLoS One 14(4):e0215622
Oettmeier C, Lee J, Döbereiner H-G (2018) Form follows function: ultrastructure of different morphotypes of Physarum polycephalum. J Phys D Appl Phys 51:134006
Ogikubo S, Nakabayashi T, Adachi T, Islam MS, Yoshizawa T, Kinjo M, Ohta N (2011) Intracellular pH sensing using autofluorescence lifetime microscopy. J Phys Chem B 115(34):10385–10390
Poulos A, Thompson GA (1971) Ether-containing lipids of the slime mold, Physarum polycephalum: II Rates of biosynthesis. Lipids 6:470–474
Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. PNAS 108(25):10190–10195
Rodrigues RM, Macko P, Palosaari T, Whelan MP (2011) Autofluorescence microscopy: a nondestructive tool to monitor mitochondrial toxicity. Toxicol Lett 206(3):281–288
Satoh H, Ueda T, Kobatake Y (1982) Primary oscillator of contractional rhythm in the plasmodium of Physarum polycephalum: role of mitochondria. Cell Struct Funct 7:275–283
Sauer H (1982) Developmental biology of Physarum (Vol. 11). CUP archive
Schaap P, Barrantes I, Minx P, Sasaki N, Anderson RW, Bénard M, Biggar KK, Buchler NE, Bundschuh R, Chen X, Fronick C, Fulton L, Golderer G, Jahn N, Knoop V, Landweber LF, Maric C, Miller D, Noegel AA, Peace R, Pierron G, Sasaki T, Schallenberg-Rüdinger M, Schleicher M, Singh R, Spaller T, Storey KB, Suzuki T, Tomlinson C, Tyson JJ, Warren WC, Werner ER, Werner-Felmayer G, Wilson RK, Winckler T, Gott JM, Glöckner G, Marwan W (2016) The Physarum polycephalum genome reveals extensive use of prokaryotic two-component and metazoan-type tyrosine kinase signaling. Genome Biol Evol 8(1):109–125
Song SB, Hwang ES (2019) A rise in ATP, ROS, and mitochondrial content upon glucose withdrawal correlates with a dysregulated mitochondria turnover mediated by the activation of the protein deacetylase SIRT1. Cells 8(1):11
Stockem W, Brix K (1994) Analysis of microfilament organization and contractile activities in Physarum. Int Rev Cytol 149:145–215
Takano H, Abe T, Sakurai R, Moriyama Y, Miyazawa Y, Nozaki H, Kawano S, Sasaki N, Kuroiwa T (2001) The complete DNA sequence of the mitochondrial genome of Physarum polycephalum. Mol Gen Genet 264:539, 545
Visser W, van Spronsen EA, Nanninga N, Pronk JT, Kuenen JG, van Dijken JP (1995) Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 67(3):243–253
Wappler EA, Institoris A, Dutta S, Katakam PVG, Busija DW (2013) Mitochondrial dynamics associated with oxygen-glucose deprivation in rat primary neuronal cultures. PLoS One 8(5):e63206
Weibel ER, Gomez DM (1962) A principle for counting tissue structures on random sections. J Appl Physiol 17:343–348
Wiesner RJ, Aschenbrenner V, Ruegg JC, Zak R (1994) Coordination of nuclear and mitochondrial gene expression during the development of cardiac hypertrophy in rats. Am J Phys 267:C229–C235
Acknowledgments
We would especially like to thank Prof. Dr. Wolfgang Marwan for kindly providing us with strains of P. polycephalum, and for comments regarding the AMPK pathway, which greatly improved the manuscript. We are grateful to Prof. Dr. Reimer Stick and Ute Helmboldt-Caesar for providing expertise and access to the TEM. We thank Anja Bammann and John Lee for experimental assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have conflict of interest.
Additional information
Handling Editor: Ralph Gräf
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Oettmeier, C., Döbereiner, HG. Mitochondrial numbers increase during glucose deprivation in the slime mold Physarum polycephalum. Protoplasma 256, 1647–1655 (2019). https://doi.org/10.1007/s00709-019-01410-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-019-01410-1