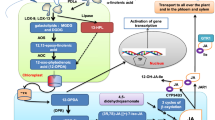
Abstract
NAM, ATAF, and CUC (NAC) domain-containing proteins are plant-specific transcription factors involved in stress responses and developmental regulation. MusaVND2 and MusaVND3 are vascular-related NAC domain-containing genes encoding for nuclear-localized proteins. The transcript level of MusaVND2 and MusaVND3 are gradually induced after induction of lignification conditions in banana embryogenic cells. Banana embryogenic cells differentiated to tracheary element-like cells after overexpression of MusaVND2 and MusaVND3 with a differentiation frequency of 63.5 and 23.4 %, respectively, after ninth day. Transgenic banana plants overexpressing either of MusaVND2 or MusaVND3 showed ectopic secondary wall deposition as well as transdifferentiation of cells into tracheary elements. Transdifferentiation to tracheary element-like cells was observed in cortical cells of corm and in epidermal and mesophyll cells of leaves of transgenic plants. Elevated levels of lignin and crystalline cellulose were detected in the transgenic banana lines than control plants. The results obtained are useful for understanding the molecular regulation of secondary wall development in banana.










Similar content being viewed by others
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Clarke WP, Radnidge P, Lai TE, Jensen PD, Hardin MT (2008) Digestion of waste bananas to generate energy in Australia. Waste Manag 28:527–533
Cote FX, Domergue R, Monmarson S, Schwendiman J, Teisson C, Escalant JV (1996) Embryogenic cell suspensions from the male flower of Musa AAA cv. Grand Nain Physiol Plant 97:285–290
Eudes A, Sathitsuksanoh N, Baidoo EE, George A, Liang Y, Yang F, Singh S, Keasling JD, Simmons BA, Loqué D (2015) Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol J. doi:10.1111/pbi.12310
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–63
Ganapathi TR, Higgs NS, Balint Kurti PJ, Arntzen CJ, May GD, Van Eck JM (2001) Agrobacterium mediated transformation of embryogenic cell suspensions of the banana cultivar Rasthali (AAB). Plant Cell Rep 20:157–162
Ganapathi TR, Sidhaa M, Suprasannaa P, Ujjappa KM, Bapat VA, D’Souza SF (2008) Field performance and RAPD analysis of gamma-irradiated variants of banana cultivar ‘Giant Cavendish’ (AAA). Int J Fruit Sci 8:147–159
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 117:808–12
Iwase A, Hideno A, Watanabe K, Mitsuda N, Ohme-Takagi M (2009) A chimeric NST repressor has the potential to improve glucose productivity from plant cell walls. J Biotechnol 142:279–84
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 16:1855–1860
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Lange BM, Lapierre C, Sandermann H Jr (1995) Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol 108:1277–1287
Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697
Ma QH (2009) The expression of cafferic acid 3-O-methyltransferase in two wheat genotypes differing in lodging resistance. J Exp Bot 60:2763–2771
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell 17:2993–3006
Nakashima J, Takabe K, Fujita M, Saiki H (1997) Inhibition of phenylalanine ammonia-lyase activity causes the depression of lignin accumulation of secondary wall thickening in isolated Zinnia mesophyll cells. Protoplasma 196:99–107
Ogita S, Nomura T, Kishimoto T, Kato Y (2012) A novel xylogenic suspension culture model for exploring lignification in Phyllostachys bamboo. Plant Methods 8:40
Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22:3461–73
Ohtani M, Nishikubo N, Xu B, Yamaguchi M, Mitsuda N, Goué N, Shi F, Ohme-Takagi M, Demura T (2011) A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J 67:499–512
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ona T, Sonoda T, Ito K, Shibata M, Tamai Y, Kojima Y, Ohshima J, Yokota S, Yoshizawa N (2001) Investigation of relationships between cell and pulp properties in Eucalyptus by examination of within-tree property variations. Wood Sci Technol 35:229–243
Ookawa T, Yasuda K, Kato H, Sakai M, Seto M et al (2010) Biomass production and lodging resistance in ‘leaf star’, a new long-culm rice forage cultivar. Plant Prod Sci 13:58–66
Plomion C, Leprovost G, Stokes A (2001) Wood formation in trees. Plant Physiol 127:1513–1523
Sakamoto S, Mitsuda N (2015) Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol 56:299–310
Schaewen AV, Langenkamper G, Graeve GK, Wenderoth I, Scheibe R (1995) Molecular characterization of the plastidic glucose-6-phosphate dehydrogenase from potato in comparison to its cytosolic counterpart. Plant Physiol 109:1327–1335
Seong ES, Yoo JH, Lee JG, Kim HY, Hwang IS, Heo K, Kim JK, Lim JD, Sacks EJ, Yu CY (2013) Antisense-overexpression of the MsCOMT gene induces changes in lignin and total phenol contents in transgenic tobacco plants. Mol Biol Rep 40:1979–86
Sewalt VJH, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb CJ, Dixon RA (1997) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol 115:41–50
Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda-Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517:571–5
Tock JY, Lai CL, Lee KT, Tan KT, Bhatia S (2010) Banana biomass as potential renewable energy resource: a Malaysian case study. Renew Sustainable Energy Rev 14:798–805
Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32:420
Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11:278–85
Velásquez-Arredondo HI, Ruiz-Colorado AA, De Oliveira S Jr (2010) Ethanol production process from banana fruit and its lignocellulosic residues: energy analysis. Energy 35:3081–3087
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664
Yamaguchi M, Demura T (2010) Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol 27:237–242
Yamaguchi M, Goué N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, Nishikubo N, Kubo M, Katayama Y, Kakegawa K, Dupree P, Demura T (2010) VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol 153:906–914
Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS, Sun L, Zheng K, Tang K, Auer M, Scheller HV, Loqué D (2013) Engineering secondary cell wall deposition in plants. Plant Biotechnol J 11:325–35
Yu Q, Li B, Nelson CD, McKeand SE, Batista VB, Mullin TJ (2006) Association of the cad-n1 allele with increased stem growth and wood density in full-sib families of loblolly pine. Tree Genet Genomes 2:98–108
Zhong R, Ye ZH (2007) Regulation of cell wall biosynthesis. Curr Opin Plant Biol 10:564–572
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Zhong R, Richardson EA, Ye ZH (2007a) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Zhong R, Lee C, Ye ZH (2010) Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 152:1044–55
Zhong R, Richardson EA, Ye ZH (2007b) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792
Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21:248–66
Zhou J, Zhong R, Ye ZH (2014) Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9, e105726
Acknowledgments
Authors thank Dr. SP Kale, Head, Nuclear Agriculture and Biotechnology Division, BARC for his constant encouragement.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Burkhard Becker
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
(PDF 169 kb)
Online resource 2
(PDF 35 kb)
Online resource 3
(PDF 4 kb)
Rights and permissions
About this article
Cite this article
Negi, S., Tak, H. & Ganapathi, T.R. Functional characterization of secondary wall deposition regulating transcription factors MusaVND2 and MusaVND3 in transgenic banana plants. Protoplasma 253, 431–446 (2016). https://doi.org/10.1007/s00709-015-0822-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-015-0822-5