Abstract
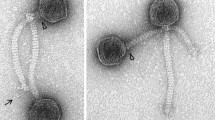
Bacillus subtilis is a Gram-positive bacterium that is widely used in fermentation and in the pharmaceutical industry. Phage contamination occasionally occurs in various fermentation processes and causes significant economic loss. Here, we report the isolation and characterization of a temperate B. subtilis phage, termed phi18-2, from spore powder manufactured in a fermentation plant. Transmission electron microscopy showed that phi18-2 has a symmetrical polyhedral head and a long noncontractile tail. Receptor analysis showed that phi18-2 recognizes wall teichoic acid (WTA) for infection. The phage virions have a linear double-stranded DNA genome of 64,467 bp with identical direct repeat sequences of 309 bp at each end of the genome. In lysogenic cells, the phage genome was found to be present in the cytoplasm without integration into the host cell chromosome, and possibly as a linear phage-plasmid with unmodified ends. Our data may provide some insight into the molecular basis of the unique lysogenic cycle of phage phi18-2.








Similar content being viewed by others
Data availability
All data supporting the conclusions of the study are included in the manuscript.
References
Abd El-Hack ME, El-Saadony MT, Shafi ME, Qattan SYA, Batiha GE, Khafaga AF, Abdel-Moneim A-ME, Alagawany M (2020) Probiotics in poultry feed: a comprehensive review. J Anim Physiol Anim Nutr 104:1835–1850
Ackermann HW (1998) Tailed bacteriophages: the order caudovirales. Adv Virus Res 51:135–201
Aliakbar Ahovan Z, Hashemi A, De Plano LM, Gholipourmalekabadi M, Seifalian A (2020) Bacteriophage based biosensors: trends, outcomes and challenges. Nanomaterials (Basel) 10:501.
Allison SE, D’Elia MA, Arar S, Monteiro MA, Brown ED (2011) Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168*. J Biol Chem 286:23708–23716
Alonso JC, Luder G, Stiege AC, Chai S, Weise F, Trautner TA (1997) The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204:201–212
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cai D, Rao Y, Zhan Y, Wang Q, Chen S (2019) Engineering Bacillus for efficient production of heterologous protein: current progress, challenge and prospect. J Appl Microbiol 126:1632–1642
Casjens SR, Hendrix RW (2015) Bacteriophage lambda: early pioneer and still relevant. Virology 479–480:310–330
Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H (2004) Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686
Chukeatirote E, Phongtang W, Kim J, Jo A, Jung LS, Ahn J (2018) Significance of bacteriophages in fermented soybeans: A review. Biomol Concepts 9:131–142
Dean DH, Orrego JC, Hutchison KW, Halvorson HO (1976) New temperate bacteriophage for Bacillus subtilis, rho 11. Journal of virology 20:509–519
Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R (2017) Communication between viruses guides lysis–lysogeny decisions. Nature 541:488–493
Errington J, Aart LTV (2020) Microbe Profile: Bacillus subtilis: model organism for cellular development, and industrial workhorse. Microbiology (Reading) 166:425–427
Gabiatti N, Yu P, Mathieu J, Lu GW, Wang X, Zhang H, Soares HM, Alvarez PJJ (2018) Bacterial endospores as phage genome carriers and protective shells. Appl Environ Microbiol 84:e01186–18.
Garneau JE, Moineau S (2011) Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb Cell Fact 10(Suppl 1):S20
Ghosh K, Kang HS, Hyun WB, Kim KP (2018) High prevalence of Bacillus subtilis-infecting bacteriophages in soybean-based fermented foods and its detrimental effects on the process and quality of Cheonggukjang. Food Microbiol 76:196–203
Gillis A, Mahillon J (2014) Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: Past, present and future. Viruses 6:2623–2672
Giraffa G, Zago M, Carminati D (2017) Lactic acid bacteria bacteriophages in dairy products: problems and solutions. In: P. Poltronieri (ed) Microbiol Dairy Process, pp 233–250.
Groth AC, Calos MP (2004) Phage integrases: biology and applications. J Mol Biol 335:667–678
Hemphill HE, Whiteley HR (1975) Bacteriophages of Bacillus subtilis. Bacteriol Rev 39:257–315
Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB (2017) Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11:1511–1520
Kordi M, Salami R, Bolouri P, Delangiz N, AsgariLajayer B, van Hullebusch ED (2022) White biotechnology and the production of bio-products. Syst Microbiol Biomanuf 2:413–429
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lazarevic V, Soldo B, Médico N, Pooley H, Bron S, Karamata D (2005) Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71:39–45
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079
Liu M, Bischoff KM, Gill JJ, Mire-Criscione MD, Berry JD, Young R, Summer EJ (2015) Bacteriophage application restores ethanol fermentation characteristics disrupted by Lactobacillus fermentum. Biotechnol Biofuels 8:132
Los M (2012) Minimization and prevention of phage infections in bioprocesses. Methods Mol Biol 834:305–315
Martin AC, Lopez R, Garcia P (1996) Analysis of the complete nucleotide sequence and functional organization of the genome of Streptococcus pneumoniae bacteriophage Cp-1. J Virol 70:3678–3687
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Meijer WJ, Castilla-Llorente V, Villar L, Murray H, Errington J, Salas M (2005) Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage phi29. EMBO J 24:3647–3657
Pfeifer E, Moura de Sousa JA, Touchon M, Rocha EPC (2021) Bacteria have numerous distinctive groups of phage-plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res 49:2655–2673
Piligrimova EG, Kazantseva OA, Kazantsev AN, Nikulin NA, Skorynina AV, Koposova ON, Shadrin AM (2021) Putative plasmid prophages of Bacillus cereus sensu lato may hold the key to undiscovered phage diversity. Sci Rep 11:7611
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28:i333–i339
Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW (2000) Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol 299:53–73
Redondo RA, Kupczok A, Stift G, Bollback JP (2013) Complete genome sequence of the novel phage MG-B1 infecting Bacillus weihenstephanensis. Genome Announc 1:e00216–13.
Rohwer F, Prangishvili D, Lindell D (2009) Roles of viruses in the environment. Environ Microbiol 11:2771–2774
Sambrook J (2001) Molecular cloning: a laboratory manual. Third edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., ©2001
Samson JE, Moineau S (2013) Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu Rev Food Sci Technol 4:347–368
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Schilling T, Hoppert M, Hertel R (2018) Genomic analysis of the recent viral isolate vB_BthP-Goe4 reveals increased diversity of φ29-Like phages. Viruses 10(11):624.
Sonenshein AL (2006) Bacteriophages: how bacterial spores capture and protect phage DNA. Current Biology 16:R14–R16
Spizizen J (1958) Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci U S A 44:1072–1078
Su Y, Liu C, Fang H, Zhang D (2020) Bacillus subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microbial Cell Factories 19:173
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010
Warner FD, Kitos GA, Romano MP, Hemphill HE (1977) Characterization of SPβ: a temperate bacteriophage from Bacillus subtilis 168M. Canadian Journal of Microbiology 23:45–51
Weigel C, Seitz H (2006) Bacteriophage replication modules. FEMS Microbiol Rev 30:321–381
Zhang Z, Liang L, Li D, Li Y, Sun Q, Li Y, Yang H (2023) Bacillus subtilis phage phi18: genomic analysis and receptor identification. Arch Virol 168:17
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31970150).
Author information
Authors and Affiliations
Contributions
Yutong Li and Hongjiang Yang designed the research. Yutong Li, Yansheng Huo, Li Liang, and Donghang Li conducted the experiments. Yutong Li analyzed data and wrote the draft. Hongjiang Yang wrote and refined the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Handling Editor: Johannes Wittmann
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Huo, Y., Liang, L. et al. Bacillus phage phi18-2 is a novel temperate virus with an unintegrated genome present in the cytoplasm of lysogenic cells as a linear phage-plasmid. Arch Virol 169, 81 (2024). https://doi.org/10.1007/s00705-024-06014-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-06014-6