Abstract
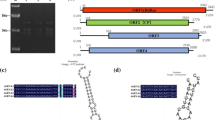
A double-stranded RNA (dsRNA) mycovirus was isolated from Talaromyces neofusisporus isolate HJ1-6 and named "Talaromyces neofusisporus chrysovirus 1" (TnCV1). It was found to consist of four dsRNA segments (TnCV1-1, TnCV1-2, TnCV1-3, and TnCV1-4) with lengths of 3595 bp, 3063 bp, 3054 bp, and 2876 bp, respectively. Sequence analysis showed that TnCV1-1 contains an open reading frame (ORF) encoding a putative RNA-dependent RNA polymerase (RdRp) of 1136 amino acids (aa), TnCV1-2 contains an ORF encoding a hypothetical protein of 906 aa, TnCV1-3 contains an ORF encoding a putative capsid protein (CP) of 938 aa, and TnCV1-4 contains an ORF encoding a hypothetical protein of 849 aa. The 5’ and 3’ untranslated regions (UTRs) of TnCV1-1, TnCV1-2, TnCV1-3, and TnCV1-4 showed a high degree of sequence similarity to each other. Phylogenetic analysis based on RdRp sequences suggested that TnCV1 is a new member of the genus Alphachrysovirus in the family Chrysoviridae. This is the first chrysovirus isolated from T. neofusisporus.


Similar content being viewed by others
References
Xie J, Jiang D (2014) New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52(1):45–68
Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D (2014) Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Pro Natl Acad Sci USA 111:12205–12210
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Pro Natl Acad Sci USA 107:8387–8392
Mu F, Li B, Cheng SI, Jia J, Jiang D, Fu Y, Cheng J, Lin Y, Chen T, Xie J (2021) Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. PLoS Pathog 17:e1009823
Krupovic M, Said AG, Jiang D, Varsani A (2016) Genomoviridae: A new family of widespread single-stranded DNA viruses. Arch Virol 161:2633–2643
Kotta-Loizou I, Castón JR, Coutts RH, Hillman BI, Jiang D, Kim D-H, Moriyama H, Suzuki N, Consortium IR (2020) ICTV virus taxonomy profile: Chrysoviridae. J Gen Virol 101:143
Luque D, Gómez-Blanco J, Garriga D, Brilot AF, González JM, Havens WM, Carrascosa JL, Trus BL, Verdaguer N, Ghabrial SA (2014) Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage. Pro Natl Acad Sci USA 111:7641–7646
McNeill J, Barrie F, Buck W, Demoulin V, Greuter We, Hawksworth D, Herendeen P, Knapp S, Marhold K, Prado J (2012) International Code of Nomenclature for algae, fungi and plants (Melbourne Code), vol 154. Koeltz Scientific Books Königstein
Wang QM, Zhang YH, Wang B, Wang L (2016) Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China. Sci Rep-UK 6:1–9
Samson RA, Visagie CM, Houbraken J, Hong S-B, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB (2005) Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 53:1–27
Morris T, Dodds J (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:855
Jiang Y, Wang J, Yang B, Wang Q, Yu W (2019) Molecular characterization of a debilitation-associated partitivirus infecting the pathogenic fungus Aspergillus flavus. Front Microbiol 10:626
Jiang Y, Zhang T, Luo C, Jiang D, Li G, Li Q, Hsiang T, Huang J (2015) Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res 195:47–56
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Funding
This study was funded by the National Natural Science Foundation of China (32000017 and 31660012), the Guizhou Provincial Natural Science Foundation [ZK(2021)zhongdian030], the Department of Education of Guizhou Province (CN) [KY(2021)313], the Regional Common Diseases and Adult Stem Cell Transformation Research and Innovation Platform of Guizhou Provincial Department of Science and Technology [(2019)4008], and Guizhou Provincial Natural Science Foundation [ZK[2022] Key Program 039].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling editor: Robert H.A. Coutts
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yinhui Jiang and Xun Tian contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, Y., Tian, X., Liu, X. et al. Complete genome sequence of a novel chrysovirus infecting Talaromyces neofusisporus. Arch Virol 167, 2789–2793 (2022). https://doi.org/10.1007/s00705-022-05582-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05582-9