Abstract
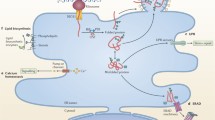
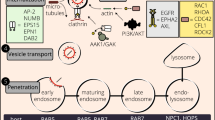
Viruses can survive only in living cells, where they depend on the host’s enzymatic system for survival and reproduction. Virus–host interactions are complex. On the one hand, hosts express host-restricted factors to protect the host cells from viral infections. On the other hand, viruses recruit certain host factors to facilitate their survival and transmission. The identification of host factors critical to viral infection is essential for comprehending the pathogenesis of contagion and developing novel antiviral therapies that specifically target the host. Receptor for activated C kinase 1 (RACK1), an evolutionarily conserved host factor that exists in various eukaryotic organisms, is a promising target for antiviral therapy. This review primarily summarizes the roles of RACK1 in regulating different viral life stages, particularly entry, replication, translation, and release.



Similar content being viewed by others
References
Giovannoni F, Bosch I, Polonio CM, Torti MF, Wheeler MA, Li Z, Romorini L, Rodriguez Varela MS, Rothhammer V, Barroso A, Tjon EC, Sanmarco LM, Takenaka MC, Modaresi SMS, Gutiérrez-Vázquez C, Zanluqui NG, Dos Santos NB, Munhoz CD, Wang Z, Damonte EB, Sherr D, Gehrke L, Peron JPS, Garcia CC, Quintana FJ (2020) AHR is a Zika virus host factor and a candidate target for antiviral therapy. Nat Neurosci 23:939–951. https://doi.org/10.1038/s41593-020-0664-0
Linero FN, Sepúlveda CS, Giovannoni F, Castilla V, García CC, Scolaro LA, Damonte EB (2012) Host cell factors as antiviral targets in arenavirus infection. Viruses 4:1569–1591. https://doi.org/10.3390/v4091569
Mochly-Rosen D, Khaner H, Lopez J (1991) Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci USA 88:3997–4000. https://doi.org/10.1073/pnas.88.9.3997
Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D (1994) Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA 91:839–843. https://doi.org/10.1073/pnas.91.3.839
Adams DR, Ron D, Kiely PA (2011) RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal 9:22. https://doi.org/10.1186/1478-811x-9-22
Sondek J, Siderovski DP (2001) Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharmacol 61:1329–1337. https://doi.org/10.1016/s0006-2952(01)00633-5
Steele MR, McCahill A, Thompson DS, MacKenzie C, Isaacs NW, Houslay MD, Bolger GB (2001) Identification of a surface on the beta-propeller protein RACK1 that interacts with the cAMP-specific phosphodiesterase PDE4D5. Cell Signal 13:507–513. https://doi.org/10.1016/s0898-6568(01)00167-x
Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko MP, Reiner O, Smith TF, Neer EJ (1996) Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry 35:13985–13994. https://doi.org/10.1021/bi9612879
Zhang X, Jain R, Li G (2016) Roles of Rack1 proteins in fungal pathogenesis. Biomed Res Int 2016:4130376. https://doi.org/10.1155/2016/4130376
Ron D, Jiang Z, Yao L, Vagts A, Diamond I, Gordon A (1999) Coordinated movement of RACK1 with activated betaIIPKC. J Biol Chem 274:27039–27046. https://doi.org/10.1074/jbc.274.38.27039
Usacheva A, Smith R, Minshall R, Baida G, Seng S, Croze E, Colamonici O (2001) The WD motif-containing protein receptor for activated protein kinase C (RACK1) is required for recruitment and activation of signal transducer and activator of transcription 1 through the type I interferon receptor. J Biol Chem 276:22948–22953. https://doi.org/10.1074/jbc.M100087200
Xu Y, Wang N, Ling F, Li P, Gao Y (2009) Receptor for activated C-kinase 1, a novel binding partner of adiponectin receptor 1. Biochem Biophys Res Commun 378:95–98. https://doi.org/10.1016/j.bbrc.2008.11.026
Vomastek T, Iwanicki MP, Schaeffer HJ, Tarcsafalvi A, Parsons JT, Weber MJ (2007) RACK1 targets the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway to link integrin engagement with focal adhesion disassembly and cell motility. Mol Cell Biol 27:8296–8305. https://doi.org/10.1128/mcb.00598-07
Zhu L, Chen W, Li G, Chen H, Liao W, Zhang L, Xiao X (2019) Upregulated RACK1 attenuates gastric cancer cell growth and epithelial-mesenchymal transition via suppressing Wnt/β-catenin signaling. Onco Targets Ther 12:4795–4805. https://doi.org/10.2147/ott.S205869
Duan F, Wu H, Jia D, Wu W, Ren S, Wang L, Song S, Guo X, Liu F, Ruan Y, Gu J (2018) O-GlcNAcylation of RACK1 promotes hepatocellular carcinogenesis. J Hepatol 68:1191–1202. https://doi.org/10.1016/j.jhep.2018.02.003
Masi M, Garattini E, Bolis M, Di Marino D, Maraccani L, Morelli E, Grolla AA, Fagiani F, Corsini E, Travelli C, Govoni S, Racchi M, Buoso E (2020) OXER1 and RACK1-associated pathway: a promising drug target for breast cancer progression. Oncogenesis 9:105. https://doi.org/10.1038/s41389-020-00291-x
Gandin V, Senft D, Topisirovic I, Ronai ZA (2013) RACK1 function in cell motility and protein synthesis. Genes Cancer 4:369–377. https://doi.org/10.1177/1947601913486348
Wang W, Wang X, Wang X, Ahmed S, Hussain S, Zhang N, Ma Y, Wang S (2019) Integration of RACK1 and ethylene signaling regulates plant growth and development in Arabidopsis. Plant Sci 280:31–40. https://doi.org/10.1016/j.plantsci.2018.11.009
Jones JE, Le Sage V, Lakdawala SS (2021) Viral and host heterogeneity and their effects on the viral life cycle. Nat Rev Microbiol 19:272–282. https://doi.org/10.1038/s41579-020-00449-9
Goodsell DS (2015) Illustrations of the HIV life cycle. Curr Top Microbiol Immunol 389:243–252. https://doi.org/10.1007/82_2015_437
Helenius A (2018) Virus entry: looking back and moving forward. J Mol Biol 430:1853–1862. https://doi.org/10.1016/j.jmb.2018.03.034
Zhang Y, Cao G, Zhu L, Chen F, Zar MS, Wang S, Hu X, Wei Y, Xue R, Gong C (2017) Integrin beta and receptor for activated protein kinase C are involved in the cell entry of Bombyx mori cypovirus. Appl Microbiol Biotechnol 101:3703–3716. https://doi.org/10.1007/s00253-017-8158-z
Takada Y, Ye X, Simon S (2007) The integrins. Genome Biol 8:215. https://doi.org/10.1186/gb-2007-8-5-215
Zhong Y, Tang X, Sheng X, Xing J, Zhan W (2019) Voltage-dependent anion channel protein 2 (VDAC2) and receptor of activated protein c kinase 1 (RACK1) act as functional receptors for lymphocystis disease virus infection. J Virol 93:e00122-19. https://doi.org/10.1128/jvi.00122-19
Sheng X, Zhong Y, Zeng J, Tang X, Xing J, Chi H, Zhan W (2020) Lymphocystis disease virus (Iridoviridae) enters flounder (Paralichthys olivaceus) gill cells via a caveolae-mediated endocytosis mechanism facilitated by viral receptors. Int J Mol Sci 21:4722. https://doi.org/10.3390/ijms21134722
Mercer J, Schelhaas M, Helenius A (2010) Virus entry by endocytosis. Annu Rev Biochem 79:803–833. https://doi.org/10.1146/annurev-biochem-060208-104626
McKenzie J, El-Guindy A (2015) Epstein–Barr virus lytic cycle reactivation. Curr Top Microbiol Immunol 391:237–261. https://doi.org/10.1007/978-3-319-22834-1_8
Baumann M, Gires O, Kolch W, Mischak H, Zeidler R, Pich D, Hammerschmidt W (2000) The PKC targeting protein RACK1 interacts with the Epstein–Barr virus activator protein BZLF1. Eur J Biochem 267:3891–3901. https://doi.org/10.1046/j.1432-1327.2000.01430.x
Nyamweya S, Hegedus A, Jaye A, Rowland-Jones S, Flanagan KL, Macallan DC (2013) Comparing HIV-1 and HIV-2 infection: lessons for viral immunopathogenesis. Rev Med Virol 23:221–240. https://doi.org/10.1002/rmv.1739
Staudt RP, Alvarado JJ, Emert-Sedlak LA, Shi H, Shu ST, Wales TE, Engen JR, Smithgall TE (2020) Structure, function, and inhibitor targeting of HIV-1 Nef-effector kinase complexes. J Biol Chem 295:15158–15171. https://doi.org/10.1074/jbc.REV120.012317
Gallina A, Rossi F, Milanesi G (2001) Rack1 binds HIV-1 Nef and can act as a Nef-protein kinase C adaptor. Virology 283:7–18. https://doi.org/10.1006/viro.2001.0855
Coronel-Ruiz C, Gutiérrez-Barbosa H, Medina-Moreno S, Velandia-Romero ML, Chua JV, Castellanos JE, Zapata JC (2020) Humanized mice in dengue research: a comparison with other mouse models. Vaccines (Basel). https://doi.org/10.3390/vaccines8010039
Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, Kümmerer BM, Delaugerre C, Roingeard P, Vidalain PO, Amara A (2017) A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep 21:3900–3913. https://doi.org/10.1016/j.celrep.2017.11.094
Subramani C, Nair VP, Anang S, Mandal SD, Pareek M, Kaushik N, Srivastava A, Saha S, Shalimar NB, Ranjith-Kumar CT, Surjit M (2018) Host–virus protein interaction network reveals the involvement of multiple host processes in the life cycle of hepatitis E virus. mSystems. https://doi.org/10.1128/mSystems.00135-17
Lee JS, Tabata K, Twu WI, Rahman MS, Kim HS, Yu JB, Jee MH, Bartenschlager R, Jang SK (2019) RACK1 mediates rewiring of intracellular networks induced by hepatitis C virus infection. PLoS Pathog 15:e1008021. https://doi.org/10.1371/journal.ppat.1008021
Zhang Y, Hou P, He DC, Wang H, He H (2021) RACK1 degrades MAVS to promote bovine ephemeral fever virus replication via upregulating E3 ubiquitin ligase STUB1. Vet Microbiol 257:109096. https://doi.org/10.1016/j.vetmic.2021.109096
Bi J, Zhao Q, Zhu L, Li X, Yang G, Liu J, Yin G (2018) RACK1 is indispensable for porcine reproductive and respiratory syndrome virus replication and NF-κB activation in Marc-145 cells. Sci Rep 8:2985. https://doi.org/10.1038/s41598-018-21460-4
Liu X, Bi J, Zhao Q, Li M, Zuo Q, Wang X, Lan R, Li X, Yang G, Liu J, Yin G (2019) Overexpression of RACK1 enhanced the replication of porcine reproductive and respiratory syndrome virus in Marc-145 cells and promoted the NF-κB activation via upregulating the expression and phosphorylation of TRAF2. Gene 709:75–83. https://doi.org/10.1016/j.gene.2019.05.046
Yang C, Lan R, Wang X, Zhao Q, Li X, Bi J, Wang J, Yang G, Lin Y, Liu J, Yin G (2020) Integrin β3, a RACK1 interacting protein, is critical for porcine reproductive and respiratory syndrome virus infection and NF-κB activation in Marc-145 cells. Virus Res 282:197956. https://doi.org/10.1016/j.virusres.2020.197956
Yang C, Zuo Q, Liu X, Zhao Q, Pu H, Gao L, Zhao L, Guo Z, Lin Y, Liu J, Bi J, Yin G (2021) Small molecule screening identified cepharanthine as an inhibitor of porcine reproductive and respiratory syndrome virus infection in vitro by suppressing integrins/ILK/RACK1/PKCα/NF-κB signalling axis. Vet Microbiol 255:109016. https://doi.org/10.1016/j.vetmic.2021.109016
Lin W, Zhang Z, Xu Z, Wang B, Li X, Cao H, Wang Y, Zheng SJ (2015) The association of receptor of activated protein kinase C 1(RACK1) with infectious bursal disease virus viral protein VP5 and voltage-dependent anion channel 2 (VDAC2) inhibits apoptosis and enhances viral replication. J Biol Chem 290:8500–8510. https://doi.org/10.1074/jbc.M114.585687
Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R (2010) Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol 17:547–554. https://doi.org/10.1038/nsmb.1810
Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, Baumert TF, Schuster C, Imler JL (2014) RACK1 controls IRES-mediated translation of viruses. Cell 159:1086–1095. https://doi.org/10.1016/j.cell.2014.10.041
Hyodo K, Suzuki N, Okuno T (2019) Hijacking a host scaffold protein, RACK1, for replication of a plant RNA virus. New Phytol 221:935–945. https://doi.org/10.1111/nph.15412
Zhang C, He L, Kang K, Chen H, Xu L, Zhang Y (2014) Screening of cellular proteins that interact with the classical swine fever virus non-structural protein 5A by yeast two-hybrid analysis. J Biosci 39:63–74. https://doi.org/10.1007/s12038-013-9411-y
Wang X, Gao L, Yang X, Zuo Q, Lan R, Li M, Yang C, Lin Y, Liu J, Yin G (2020) Porcine RACK1 negatively regulates the infection of classical swine fever virus and the NF-κB activation in PK-15 cells. Vet Microbiol 246:108711. https://doi.org/10.1016/j.vetmic.2020.108711
Gallo S, Ricciardi S, Manfrini N, Pesce E, Oliveto S, Calamita P, Mancino M, Maffioli E, Moro M, Crosti M, Berno V, Bombaci M, Tedeschi G, Biffo S (2018) RACK1 specifically regulates translation through its binding to ribosomes. Mol Cell Biol. https://doi.org/10.1128/mcb.00230-18
Miluzio A, Beugnet A, Volta V, Biffo S (2009) Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep 10:459–465. https://doi.org/10.1038/embor.2009.70
Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Marchisio PC, Biffo S (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426:579–584. https://doi.org/10.1038/nature02160
Yang Y, Wang Z (2019) IRES-mediated cap-independent translation, a path leading to hidden proteome. J Mol Cell Biol 11:911–919. https://doi.org/10.1093/jmcb/mjz091
Gray NK, Wickens M (1998) Control of translation initiation in animals. Annu Rev Cell Dev Biol 14:399–458. https://doi.org/10.1146/annurev.cellbio.14.1.399
Montero H, García-Román R, Mora SI (2015) eIF4E as a control target for viruses. Viruses 7:739–750. https://doi.org/10.3390/v7020739
Martins N, Imler JL, Meignin C (2016) Discovery of novel targets for antivirals: learning from flies. Curr Opin Virol 20:64–70. https://doi.org/10.1016/j.coviro.2016.09.005
Mokrejs M, Vopálenský V, Kolenaty O, Masek T, Feketová Z, Sekyrová P, Skaloudová B, Kríz V, Pospísek M (2006) IRESite: the database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res 34:D125-130. https://doi.org/10.1093/nar/gkj081
Johnson AG, Grosely R, Petrov AN, Puglisi JD (2017) Dynamics of IRES-mediated translation. Philos Trans R Soc Lond B Biol Sci. https://doi.org/10.1098/rstb.2016.0177
Kerr CH, Wang QS, Keatings K, Khong A, Allan D, Yip CK, Foster LJ, Jan E (2015) The 5’ untranslated region of a novel infectious molecular clone of the dicistrovirus cricket paralysis virus modulates infection. J Virol 89:5919–5934. https://doi.org/10.1128/jvi.00463-15
Hertz MI, Thompson SR (2011) Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology 411:355–361. https://doi.org/10.1016/j.virol.2011.01.005
Sun C, Querol-Audí J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH (2013) Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res 41:7512–7521. https://doi.org/10.1093/nar/gkt510
LaFontaine E, Miller CM, Permaul N, Martin ET, Fuchs G (2020) Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 545:53–62. https://doi.org/10.1016/j.virol.2020.03.004
Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, Walsh D (2017) Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546:651–655. https://doi.org/10.1038/nature22814
Ullah H, Hou W, Dakshanamurthy S, Tang Q (2019) Host targeted antiviral (HTA): functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget 10:3209–3226. https://doi.org/10.18632/oncotarget.26907
Demirov D, Gabriel G, Schneider C, Hohenberg H, Ludwig S (2012) Interaction of influenza A virus matrix protein with RACK1 is required for virus release. Cell Microbiol 14:774–789. https://doi.org/10.1111/j.1462-5822.2012.01759.x
Kim HS, Lee K, Kim SJ, Cho S, Shin HJ, Kim C, Kim JS (2018) Arrayed CRISPR screen with image-based assay reliably uncovers host genes required for coxsackievirus infection. Genome Res 28:859–868. https://doi.org/10.1101/gr.230250.117
Nielsen MH, Flygaard RK, Jenner LB (2017) Structural analysis of ribosomal RACK1 and its role in translational control. Cell Signal 35:272–281. https://doi.org/10.1016/j.cellsig.2017.01.026
Kubota T, Yokosawa N, Yokota S, Fujii N (2002) Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J Virol 76:12676–12682. https://doi.org/10.1128/jvi.76.24.12676-12682.2002
Xie T, Chen T, Li C, Wang W, Cao L, Rao H, Yang Q, Shu HB, Xu LG (2019) RACK1 attenuates RLR antiviral signaling by targeting VISA-TRAF complexes. Biochem Biophys Res Commun 508:667–674. https://doi.org/10.1016/j.bbrc.2018.11.203
Tardif M, Savard M, Flamand L, Gosselin J (2002) Impaired protein kinase C activation/translocation in Epstein–Barr virus-infected monocytes. J Biol Chem 277:24148–24154. https://doi.org/10.1074/jbc.M109036200
Qin C, Niu C, Shen Z, Zhang Y, Liu G, Hou C, Dong J, Zhao M, Cheng Q, Yang X, Zhang J (2021) RACK1 T50 phosphorylation by AMPK potentiates its binding with IRF3/7 and inhibition of type 1 IFN production. J Immunol (Baltimore, MD, 1950) 207:1411–1418. https://doi.org/10.4049/jimmunol.2100086
Funding
This research was support by the National Natural Science Foundation of China (grant no. 81971945).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Data collection and analysis were performed by Yuhan Li, Qingru Yang, Lulu Wang, and Xiaolan Liu. The first draft of the manuscript was written by Yan Wang and Xiaorong Qiao. Hua Wang and Hongxing Shen commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Qiao, X., Li, Y. et al. Role of the receptor for activated C kinase 1 during viral infection. Arch Virol 167, 1915–1924 (2022). https://doi.org/10.1007/s00705-022-05484-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05484-w