Abstract
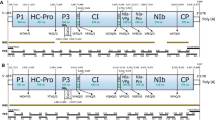
A macluravirus, tentatively named alpinia oxyphylla mosaic virus (AloMV), was identified in Alpinia oxyphylla, and its complete genomic sequence determined. The positively single-stranded RNA genome is comprised of 8213 nucleotides excluding the poly (A) tail, and contains one large open reading frame encoding a polyprotein of 2,626 amino acids. Blastp search showed that the polyprotein of AloMV shared 48%~68% aa sequence identities with other reported macluraviruses. Phylogenetic analysis based on the nucleotide sequence of the polyprotein showed that AloMV, together with all other macluraviruses, clustered into the same group most closely related to cardamom mosaic virus, sharing 66.3% nt and 68% aa sequence identities, respectively. These data above suggest that AloMV represents an isolate of a putative new member within the genus Macluravirus.

Similar content being viewed by others
References
Liou R, Yan H, Hong JL (2003) Molecular evidence that aphid-transmitted Alpinia mosaic virus is a tentative member of the genus Macluravirus. Arch Virol 148:1211–1218
Gonsalves D, Trujillo E, Hoch HC (1986) Purification and some properties of a virus associated with cardamom mosaic, a new member of the Potyvirus group. Plant Dis 70:65–69
Mandal B, Vijayanandraj S, Shilpi S, Pun KB, Singh V, Pant RP, Jain RK, Varadarasan S, Varma A (2012) Disease distribution and characterisation of a new macluravirus associated with chirke disease of large cardamom. Ann Appl Biol 160:225–236
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Chung BY, Miller WA, Atkins JF, Firth AE (2008) An over-lapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105:5897–5902
Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE (2015) Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004
Wen RH, Hajimorad MR (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400:1–7
Gao F, Shen J, Liao F, Cai W, Lin S, Yang H, Chen S (2018) The first complete genome sequence of narcissus latent virus from Narcissus. Arch Virol 163:1383–1386
Zhang P, Peng J, Guo H, Chen J, Chen S, Wang J (2016) Complete genome sequence of yam chlorotic necrotic mosaic virus from Dioscorea parviflora. Arch Virol 161:1715–1717
Adams MJ, Antoniw JF, Fauquet CM (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol 150:459–479
Acknowledgements
The authors thank Dr. Xiaofei Cheng (Northeast Agricultural University) for assistance in analysis of sRNA data, two anonymous reviewers for constructive comments to improve our manuscript, and the TAX-editor for language check.
Funding
This study was financially funded by Hainan Provincial Natural Science Foundation of China (Grant no. 318MS006) and the Scientific Research Foundation for Advanced Talents, Hainan University (Grant no. KYQD(ZR)1734).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, W., Li, Z., Wang, X. et al. Complete genomic sequence of a novel macluravirus, alpinia oxyphylla mosaic virus (AloMV), identified in Alpinia oxyphylla. Arch Virol 163, 2579–2582 (2018). https://doi.org/10.1007/s00705-018-3879-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3879-6