Abstract
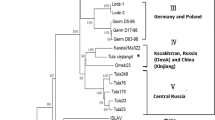
Puumala virus (PUUV) represents one of the most important hantaviruses in Central Europe. Phylogenetic analyses of PUUV strains indicate a strong genetic structuring of this hantavirus. Recently, PUUV sequences were identified in the natural reservoir, the bank vole (Myodes glareolus), collected in the northern part of Poland. The objective of this study was to evaluate the presence of PUUV in bank voles from southern Poland. A total of 72 bank voles were trapped in 2009 at six sites in this part of Poland. RT-PCR and IgG-ELISA analyses detected three PUUV positive voles at one trapping site. The PUUV-infected animals were identified by cytochrome b gene analysis to belong to the Carpathian and Eastern evolutionary lineages of bank vole. The novel PUUV S, M and L segment nucleotide sequences showed the closest similarity to sequences of the Russian PUUV lineage from Latvia, but were highly divergent to those previously found in northern Poland, Slovakia and Austria. In conclusion, the detection of a highly divergent PUUV lineage in southern Poland indicates the necessity of further bank vole monitoring in this region allowing rational public health measures to prevent human infections.




Similar content being viewed by others
References
Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB (2011) Family Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Inc., San Diego, USA, pp 725–741
Krüger DH, Schönrich G, Klempa B (2011) Human pathogenic hantaviruses and prevention of infection. Hum Vaccin 7(6):685–693
Schönrich G, Rang A, Lütteke N, Raftery MJ, Charbonnel N, Ulrich RG (2008) Hantavirus-induced immunity in rodent reservoirs and humans. Immunol Rev 225:163–189
Spitzenberger F (1999) Clethrionomys glareolus. In: Mitchell-Jones AJ (ed) The atlas of European mammals. T & AD Poyser LTD, London, pp 212–213
Heyman P, Ceianu CS, Christova I, Tordo N, Beersma M, João Alves M, Lundkvist Å, Hukic M, Papa A, Tenorio A, Zelená H, Essbauer S, Visontai I, Golovljova I, Connell J, Nicoletti L, Van Esbroeck M, Gjeruldsen Dudman S, Aberle SW, Avšič-Županc T, Korukluoglu G, Nowakowska A, Klempa B, Ulrich RG, Bino S, Engler O, Opp M, Vaheri A (2011) A five-year perspective on the situation of haemorrhagic fever with renal syndrome and status of the hantavirus reservoirs in Europe, 2005-2010. Euro Surveill 16(36):977–986
Reil D, Imholt C, Eccard JA, Jacob J (2015) Beech fructification and bank vole population dynamics - combined analyses of promoters of human Puumala Virus infections in Germany. PLoS One 10(7):e0134124
Ettinger J, Hofmann J, Enders M, Tewald F, Oehme RM, Rosenfeld UM, Ali HS, Schlegel M, Essbauer S, Osterberg A, Jacob J, Reil D, Klempa B, Ulrich RG, Krüger DH (2012) Multiple synchronous outbreaks of Puumala virus, Germany, 2010. Emerg Infect Dis 18(9):1461–1464
Weber de Melo V, Sheikh Ali H, Freise J, Kühnert D, Essbauer S, Mertens M, Wanka KM, Drewes S, Ulrich RG, Heckel G (2015) Spatiotemporal dynamics of Puumala hantavirus associated with its rodent host, Myodes glareolus. Evol Appl 8(6):545–559
Sironen T, Plyusnin A (2011) Genetics and Evolution of Hantaviruses. In: Plyusnin A, Elliot RM (eds) Bunyaviridae: molecular and cellular biology. Caister Academic Press, Norfolk, pp 61–94
Ali HS, Drewes S, Weber de Melo V, Schlegel M, Freise J, Groschup MH, Heckel G, Ulrich RG (2015) Complete genome of a Puumala virus strain from Central Europe. Virus Genes 50(2):292–298
Jääskeläinen KM, Kaukinen P, Minskaya ES, Plyusnina A, Vapalahti O, Elliott RM, Weber F, Vaheri A, Plyusnin A (2007) Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J Med Virol 79(10):1527–1536
Schlegel M, Jacob J, Krüger DH, Rang A, Ulrich RG (2014) Hantavirus emergence in rodents, insectivores and bats. In: Johnson N (ed) Role of animals in emerging viral diseases. Academic Press Waltham, MA, pp 235–291
Nowakowska A, Heyman P, Knap JP, Burzyński W, Witas M (2009) The first established focus of hantavirus infection in Poland, 2007. Ann Agric Environ Med 16(1):79–85
Michalski A, Niemcewicz M, Bielawska-Drózd A, Nowakowska A, Gaweł J, Pitucha G, Joniec J, Zielonka K, Marciniak-Niemcewicz A, Kocik J (2014) Surveillance of hantaviruses in Poland: a study of animal reservoirs and human hantavirus disease in Subcarpathia. Vector Borne Zoonotic Dis 14(7):514–522
Song JW, Baek LJ, Song KJ, Skrok A, Markowski J, Bratosiewicz-Wasik J, Kordek R, Liberski PP, Yanagihara R (2004) Characterization of Tula virus from common voles (Microtus arvalis) in Poland: evidence for geographic-specific phylogenetic clustering. Virus Genes 29(2):239–247
Gu SH, Markowski J, Kang HJ, Hejduk J, Sikorska B, Liberski PP, Yanagihara R (2013) Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol J 10:160
Gu SH, Hejduk J, Markowski J, Kang HJ, Markowski M, Połatyńska M, Sikorska B, Liberski PP, Yanagihara R (2014) Co-circulation of soricid- and talpid-borne hantaviruses in Poland. Infect Genet Evol 28:296–303
Ali HS, Drewes S, Sadowska ET, Mikowska M, Groschup MH, Heckel G, Koteja P, Ulrich RG (2014) First molecular evidence for Puumala hantavirus in Poland. Viruses 6(1):340–353
Gut AK, Gut R, Pencuła M, Jarosz MJ (2013) New cases of suspected HFRS (Hantavirus infection) in south-eastern Poland. Ann Agric Environ Med 20(3):544–548
Mertens M, Kindler E, Emmerich P, Esser J, Wagner-Wiening C, Wölfel R, Petraityte-Burneikiene R, Schmidt-Chanasit J, Zvirbliene A, Groschup MH, Dobler G, Pfeffer M, Heckel G, Ulrich RG, Essbauer SS (2011) Phylogenetic analysis of Puumala virus subtype Bavaria, characterization and diagnostic use of its recombinant nucleocapsid protein. Virus Genes 43(2):177–191
Essbauer S, Schmidt J, Conraths FJ, Friedrich R, Koch J, Hautmann W, Pfeffer M, Wölfel R, Finke J, Dobler G, Ulrich R (2006) A new Puumala hantavirus subtype in rodents associated with an outbreak of Nephropathia epidemica in South-East Germany in 2004. Epidemiol Infect 134(6):1333–1344
Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Denys C, Koivogui L, ter Meulen J, Krüger DH (2006) Hantavirus in African wood mouse, Guinea. Emerg Infect Dis 12(5):838–840
Razzauti M, Plyusnina A, Henttonen H, Plyusnin A (2008) Accumulation of point mutations and reassortment of genomic RNA segments are involved in the microevolution of Puumala hantavirus in a bank vole (Myodes glareolus) population. J Gen Virol 89(Pt 7):1649–1660
Schlegel M, Ali HS, Stieger N, Groschup MH, Wolf R, Ulrich RG (2012) Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem Genet 50(5–6):440–447
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE), 2010. New Orleans, pp 1–8
Kotlík P, Deffontaine V, Mascheretti S, Zima J, Michaux JR, Searle JB (2006) A northern glacial refugium for bank voles (Clethrionomys glareolus). Proc Natl Acad Sci USA 103(40):14860–14864
Deffontaine V, Libois R, Kotlík P, Sommer R, Nieberding C, Paradis E, Searle JB, Michaux JR (2005) Beyond the Mediterranean peninsulas: evidence of central European glacial refugia for a temperate forest mammal species, the bank vole (Clethrionomys glareolus). Mol Ecol 14(6):1727–1739
Wójcik JM, Kawałko A, Marková S, Searle JB, Kotlík P (2010) Phylogeographic signatures of northward post-glacial colonization from high-latitude refugia: a case study of bank voles using museum specimens. J Zool 281(4):249–262
Boratyński Z, Koteja P (2009) The association between body mass, metabolic rates and survival of bank voles. Functional Ecology 23(2):330–339
Faber M, Wollny T, Schlegel M, Wanka KM, Thiel J, Frank C, Rimek D, Ulrich RG, Stark K (2013) Puumala virus outbreak in Western Thuringia, Germany, 2010: epidemiology and strain identification. Zoonoses Public Health 60(8):549–554
Drewes S, Ali HS, Saxenhofer M, Rosenfeld UM, Binder F, Cuypers F, Schlegel M, Röhrs S, Heckel G, Ulrich RG (2017) Host-associated absence of human Puumala virus infections in northern and eastern Germany. Emerg Infect Dis 23(1):83–86
Razzauti M, Plyusnina A, Niemimaa J, Henttonen H, Plyusnin A (2012) Co-circulation of two Puumala hantavirus lineages in Latvia: a Russian lineage described previously and a novel Latvian lineage. J Med Virol 84(2):314–318
Straková P, Jagdmann S, Balčiauskas L, Balčiauskienė L, Drewes S, Ulrich RG (2017) Molecular detection of Puumala virus in bank voles (Myodes glareolus). Lithuania. Emerg Infect Dis 23(1):158–160
Schmidt S, Saxenhofer M, Drewes S, Schlegel M, Wanka KM, Frank R, Klimpel S, von Blanckenhagen F, Maaz D, Herden C, Freise J, Wolf R, Stubbe M, Borkenhagen P, Ansorge H, Eccard JA, Lang J, Jourdain E, Jacob J, Marianneau P, Heckel G, Ulrich RG (2016) High genetic structuring of Tula hantavirus. Arch Virol 161(5):1135–1149
Monroe MC, Morzunov SP, Johnson AM, Bowen MD, Artsob H, Yates T, Peters CJ, Rollin PE, Ksiazek TG, Nichol ST (1999) Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg Infect Dis 5(1):75–86
Plyusnin A, Morzunov SP (2001) Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol 256:47–75
Morzunov SP, Rowe JE, Ksiazek TG, Peters CJ, St Jeor SC, Nichol ST (1998) Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol 72(1):57–64
Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Mäkelä S, Mustonen J (2013) Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol 11(8):539–550
Acknowledgements
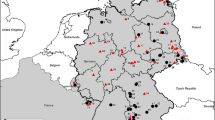
We thank Renata Świergosz-Kowalewska for help with planning the field work, Barbara Magłysz, Aneta Gaura and Adam Dzida for help in trappings, Christian Imholt, Christin Trapp, Stefan Fischer and Dörte Kaufmann for their excellent technical assistance, Nicole Reimer and Christoph Staubach for preparation of Fig. 1, and Małgorzata Sadkowska-Todys for comments on the manuscript. The work in the laboratory of RGU was supported by the Deutsche Forschungsgemeinschaft (SPP 1596 „Ecology and Species Barriers in Emerging Viral Diseases“, UL 405/1-1 to RGU) and the EU grant FP7-261504 EDENext and is catalogued by the EDENext Steering Committee as EDENext367 (http://www.edenext.eu). The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. GH was supported by Grant 31003A-149585 from the Swiss National Science Foundation. The field work was financed by FRISC project (Polish Ministry of Science, the EEA/Norwegian Financial Mechanism), and the Jagiellonian University (DS/WBINOZ/INOS/758 & 757).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Statement on the welfare of animals
All procedures on animals were approved by the First Local Bioethical Committee in Kraków (decision # 48/2007).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rosenfeld, U.M., Drewes, S., Ali, H.S. et al. A highly divergent Puumala virus lineage in southern Poland. Arch Virol 162, 1177–1185 (2017). https://doi.org/10.1007/s00705-016-3200-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3200-5