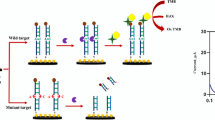
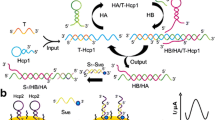
Abstract
An improved electrochemical sensor has been developed for sensitive detection of the p53 gene based on exponential amplification reaction (EXPAR) and CRISPR/Cas12a. Restriction endonuclease BstNI is introduced to specifically identify and cleave the p53 gene, generating primers to trigger the EXPAR cascade amplification. A large number of amplified products are then obtained to enable the lateral cleavage activity of CRISPR/Cas12a. For electrochemical detection, the amplified product activates Cas12a to digest the designed block probe, which allows the signal probe to be captured by the reduced graphene oxide-modified electrode (GCE/RGO), resulting in an enhanced electrochemical signal. Notably, the signal probe is labeled with large amounts of methylene blue (MB). Compared with traditional endpoint decoration, the special signal probe effectively amplifies the electrochemical signals by a factor of about 15. Experimental results show that the electrochemical sensor exhibits wide ranges from 500 aM to 10 pM and 10 pM to 1 nM, as well as a relatively low limit detection of 0.39 fM, which is about an order of magnitude lower than that of fluorescence detection. Moreover, the proposed sensor shows reliable application capability in real human serum, indicating that this work has great prospects for the construction of a CRISPR-based ultra-sensitive detection platform.
Graphical Abstract







Similar content being viewed by others
References
Levine AJ, Finlay CA, Hinds PW (2004) P53 is a tumor suppressor gene. Cell 116:S67–S69
Sigal A, Rotter V (2000) Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Can Res 60:6788–6893
Sammons MA, Nguyen T-AT, McDade SS, Fischer M (2020) Tumor suppressor p53: from engaging DNA to target gene regulation. Nucleic Acids Res 48:8848–8869
Esteban-Fernandez de Avila B, Araque E, Campuzano S, Pedrero M, Dalkiran B, Barderas R et al (2015) Dual functional graphene derivative-based electrochemical platforms for detection of the TP53 gene with single nucleotide polymorphism selectivity in biological samples. Anal Chem 87:2290–2298
Zhang C, Liu J, Xu D, Zhang T, Hu W, Feng Z (2020) Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol 12:674–687
Syahruddin E, Zaini J, Sembiring R, Baginta R, Fadhillah MR, Noor DR (2022) TP53 and EGFR mutational status in thymoma: a genetic sequencing study. Asian Pac J Cancer Prev: APJCP 23:109–114
Sadia H, Bhinder MA, Irshad A, Zahid B, Ahmed R, Ashiq S et al (2020) Determination of expression profile of p53 gene in different grades of breast cancer tissues by real time PCR. Afr Health Sci 20:1273–1282
Frazzi R, Bizzarri V, Albertazzi L, Cusenza VY, Coppolecchia L, Luminari S et al (2020) Droplet digital PCR is a sensitive tool for the detection of TP53 deletions and point mutations in chronic lymphocytic leukaemia. Br J Haematol 189:E49–E52
Fu R, Wang Y, L Y, Liu H, Zhao Q, Zhang Y, Wang C, Li Z, Jiao B, He Y (2022) CRISPR-Cas12a based fluorescence assay for organophosphorus pesticides in agricultural products. Food Chem 387:132919
Liu Y, Wang Y, Ma L, Fu R, Liu H, Cui Y, Zhao Q, Zhang Y, Jiao B, He Y (2022) A CRISPR/Cas12a-based photothermal platform for the portable detection of citrus-associated Alternaria genes using a thermometer. Int J Biol Macromol 222:2661–2669
Chertow DS (2018) Next-generation diagnostics with CRISPR. Sci 360:381–382
Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F (2019) SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 14:2986–3012
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM et al (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Sci 360:436
Li L, Li S, Wu N, Wu J, Wang G, Zhao G et al (2019) HOLMESv2: AaCRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol 8:2228–2237
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Sci 360:439
Chen JS (2021) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Sci 371:791
Wu H, Chen Y, Yang Q, Peng C, Wang X, Zhang M et al (2021) A reversible valve-assisted chip coupling with integrated sample treatment and CRISPR/Cas12a for visual detection of Vibrio parahaemolyticus. Biosens Bioelectron 188(15):113352
Yue S, Li Y, Qiao Z, Song W, Bi S (2021) Rolling circle replication for biosensing, bioimaging, and biomedicine. Trends Biotechnol 39:1160–1172
Zhou B, Lin L, Li B (2021) Exponential amplification reaction-based fluorescent sensor for the sensitive detection of tumor biomarker flap endonuclease 1. Sensors Actuators B-Chem 346:130457
Bi S, Yue S, Zhang S (2017) Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem Soc Rev 46:4281–4298
Glökler J, Lim T, Ida J, Frohme M (2021) Isothermal amplifications-a comprehensive review on current methods. Crit Rev Biochem Mol Biol 56(6):543–586
Wu H, Wu J, Liu Y, Wang H, Zou P (2019) Fluorometric determination of microRNA using arched probe-mediated isothermal exponential amplification combined with DNA-templated silver nanoclusters. Microchim Acta 186:715
Carter JG, Iturbe LO, Duprey J-LHA, Carter IR, Southern CD, Rana M et al (2021) Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc Natl Acad Sci U S A 118:e2100347118
Qing M, Chen S, Sun Z, Fan Y, Luo H, Li N (2021) Universal and programmable rolling circle amplification-CRISPR/Cas12a-mediated immobilization-free electrochemical biosensor. Anal Chem 93:7499–7507
Dai YF, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu CC (2019) Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chem Int Ed 58:17399–17405
Suea-Ngam A, Howes PD, DeMello AJ (2021) An amplification-free ultra-sensitive electrochemical CRISPR/Cas biosensor for drug-resistant bacteria detection. Chem Sci 12:12733–12743
Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W (2019) CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater 31:1905311
Zhao L, Pan H, Zhang X, Zhou Y (2020) Ultrasensitive detection of microRNA based on a homogeneous label-free electrochemical platform using G-triplex/methylene blue as a signal generator. Anal Chim Acta 1116:62–69
Wang L, Chen W, Wang Y, Zeng L, Chen T, Chen G, Chen J (2020) Ultrasensitive detection of microRNA based on a homogeneous label-free electrochemical platform using G-triplex/methylene blue as a signal generator. Biosens Bioelectron 169:112555
Qing M, Sun Z, Wang L, Du SZ, Zhou J, Tang Q et al (2021) CRISPR/Cas12a-regulated homogeneous electrochemical aptasensor for amplified detection of protein. Sensors Actuators B-Chem 348:130713
Zhou S, Sun H, Wang X, Lu P, Huo D, Li J et al (2021) Fe-hemin-metal organic frameworks/three-dimensional graphene composites with efficient peroxidase-like bioactivity for real-time electrochemical detection of extracellular hydrogen peroxide. J Electrochem Soc 168:127501
Zheng J, Zhao P, Zhou S, Chen S, Liang Y, Tian F, Zhou J et al (2019) Development of Au-Pd@UiO-66-on-ZIF-L/CC as a self-supported electrochemical sensor for in situ monitoring of cellular hydrogen peroxide. J Mater Chem B 31:1905311
Funding
This work was supported by the National Natural Science Foundation of China (No. 81772290), Chongqing science and technology commission (cstc2021jcyj-msxmX0608), the Graduate Scientific Research and Innovation Foundation of Chongqing, China (CYB22072), the Fundamental Research Funds for the Central Universities (2021CDJYGRH006), the Sichuan Science and Technology Program (2022YFSY0013), the Chongqing Graduate Tutor Team Construction Project, and the Analytical and Testing Center of Chongqing University (SEM/Raman) and the sharing fund of Chongqing University’s large equipment.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, S., Deng, L., Dong, J. et al. Electrochemical detection of the p53 gene using exponential amplification reaction (EXPAR) and CRISPR/Cas12a reactions. Microchim Acta 190, 113 (2023). https://doi.org/10.1007/s00604-023-05642-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05642-0