Abstract
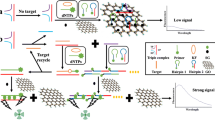
A graphene-based bioassay is described for the fluorometric determination of agrD gene transcription (mRNA) in methicillin-resistant Staphylococcus aureus (MRSA). This method includes exonuclease III (Exo III)-assisted target recycling and DNA walker cascade amplification. Hairpin1 (HP1) consists of a capture probe (CP) and DNA walker sequence. In the absence of the target, 5′-amino modified hairpin2 (HP2) labeled with carboxyfluorescein (FAM) at its 3′ terminus is covalently linked to graphene via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide (EDC/NHS) catalysis, resulting in the quenching of the FAM signal. The stem-loop structure of HP1 opens when the target is added to form partially complementary DNA/RNA hybrids. Exo III then initiates the target recycling process by cleaving the CP and DNA walker cascade reaction by automatic walking. This iterative reaction causes the FAM to dissociate from the graphene, and the fluorescence can be measured at excitation/emission wavelengths of 480/514 nm. Therefore, the target can be assayed by fluorescence. This method has a linear relationship with the concentration of target within the range 1 fM to 100 pM with a detection limit of 1 fM. The developed bioassay was used to monitor biofilm formation and investigate the mechanism of drug action with satisfactory results.
Graphical abstract

Schematic representation of the graphene-based fluorescent bioassay for agrD gene transcription in methicillin-resistant Staphylococcus aureus by using exonuclease III-aided target recycling and DNA walker cascade amplification





Similar content being viewed by others
References
Xia J, Gao J, Kokudo N, Hasegawa K, Tang W (2013) Methicillin-resistant Staphylococcus aureus antibiotic resistance and virulence. Biosci Trends 7(3):113–121
Davis JS, Van Hal S, Tong SY (2015) Combination antibiotic treatment of serious methicillin-resistant Staphylococcus aureus infections. Semin Respir Crit Care Med 36(1):3–16
Sowash MG, Uhlemann AC (2014) Community-associated methicillin-resistant Staphylococcus aureus case studies. Methods Mol Biol 1085:25–69
Tsouklidis N, Kumar R, Heindl SE, Soni R, Khan S (2020) Understanding the fight against resistance: hospital-acquired methicillin-resistant Staphylococcus aureus vs. community-acquired methicillin-resistant Staphylococcus aureus. Cureus 12(6):e8867
Oyama T, Miyazaki M, Yoshimura M, Takata T, Ohjimi H, Jimi S (2016) Biofilm-forming methicillin-resistant Staphylococcus aureus survive in Kupffer cells and exhibit high virulence in mice. Toxins (Basel) 8(7):198
Kai K (2016) Bacterial quorum sensing in symbiotic and pathogenic relationships with hosts. Biosci Biotechnol Biochem 82(3):363–371
Hawver LA, Jung SA, Ng WL (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40(5):738–752
Tahmasebi H, Dehbashi S, Arabestani MR (2019) Association between the accessory gene regulator (agr) locus and the presence of superantigen genes in clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res Notes 12(1):130
Thoendel M, Horswill AR (2009) Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem 284(33):21828–21838
Blevins T (2017) Northern blotting techniques for small RNAs. Methods Mol Biol 1456:141–162
Wada M, Lkhagvadorj E, Bian L, Wang C, Chiba Y, Nagata S, Shimizu T, Yamashiro Y, Asahara T, Nomoto K (2010) Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Appl Microbiol 108(3):779–788
Tjong V, Yu H, Hucknall A, Chilkoti A (2013) Direct fluorescence detection of RNA on microarrays by surface-initiated enzymatic polymerization. Anal Chem 85(1):426–433
Wang G, Fu Y, Ren Z, Huang J, Best S, Li X, Han G (2018) Upconversion nanocrystal ‘armoured’ silica fibres with superior photoluminescence for miRNA detection. Chem Commun (Camb) 54(49):6324–6327
Wang Y, Fan X, Yin HS, Zhou YL, Yang Y, Ai SY (2020) Photoelectrochemical immunosensor for methylated RNA detection based on WS2 and poly(U) polymerase-triggered signal amplification. Microchim Acta 187(11):596
Zhang H, Liu Y, Fu X, Yuan LH, Zhu ZJ (2015) Microfluidic bead-based assay for microRNAs using quantum dots as labels and enzymatic amplification. Microchim Acta 182(3–4):661–669
Wang Q, Liu RJ, Yang XH, Wang KM, Zhu JQ, He LL, Li Q (2016) Surface plasmon resonance biosensor for enzyme-free amplified microRNA detection based on gold nanoparticles and DNA supersandwich. Sensors Actuators B Chem 223:613–620
Jin L, Qiao J, Chen J, Xu N, Wu M (2020) Combination of area controllable sensing surface and bipolar electrode-electrochemiluminescence approach for the detection of tetracycline. Talanta 208:120404
Sun Y, Wang Y, Lau C, Chen G, Lu J (2018) Hybridization-initiated exonuclease resistance strategy for simultaneous detection of multiple microRNAs. Talanta 190:248–254
Ning Y, Chen SQ, Hu J, Li L, Cheng LJ, Lu FG (2020) Fluorometric determination of agrA gene transcription in methicillin-resistant Staphylococcus aureus with a graphene oxide-based assay using strand-displacement polymerization recycling and hybridization chain reaction. Microchim Acta 187(7):372
Tang Y, Liu M, Zhao Z, Li Q, Liang X, Tian J, Zhao S (2019) Fluorometric determination of microRNA-122 by using ExoIII-aided recycling amplification and polythymine induced formation of copper nanoparticles. Microchim Acta 186(3):133
Xu M, Gao Z, Wei Q, Chen G, Tang D (2016) Label-free hairpin DNA-scaffolded silver nanoclusters for fluorescent detection of Hg2+ using exonuclease III-assisted target recycling amplification. Biosens Bioelectron 79:411–415
Bao T, Shu H, Wen W, Zhang X, Wang S (2015) A sensitive electrochemical aptasensor for ATP detection based on exonuclease III-assisted signal amplification strategy. Anal Chim Acta 862:64–69
Pei Q, Song X, Liu S, Wang J, Leng X, Cui X, Yu J, Wang Y, Huang J (2019) A facile signal-on electrochemical DNA sensing platform for ultrasensitive detection of pathogenic bacteria based on Exo III-assisted autonomous multiple-cycle amplification. Analyst 144(9):3023–3029
Chen J, Luo ZW, Sun CJ, Huang ZJ, Zhou C, Yin S, Duan YX, Li YX (2019) Research progress of DNA walker and its recent applications in biosensor. Trends Anal Chem 120:115626
Wang DF, Vietz C, Schröder T, Acuna G, Lalkens B, Tinnefeld P (2017) A DNA walker as a fluorescence signal amplifier. Nano Lett 17(9):5368–5374
Škugor M, Valero J, Murayama K, Centola M, Asanuma H, Famulok M (2019) Orthogonally photocontrolled non-autonomous DNA walker. Angew Chem Int Ed Eng 58(21):6948–6951
Omabegho T, Sha RJ, Seeman NC (2009) A bipedal DNA Brownian motor with coordinated legs. Science 324(5923):67–71
Zhou H, Duan SY, Huang J, He FJ (2020) Ultrasensitive electrochemical biosensor for Pseudomonas aeruginosa assay based on rolling circle amplification-assisted multipedal DNA walker. Chem Commun (Camb) 56(46):6273–6276
Li D, Luo ZW, An HF, Yang EL, Wu MF, Huang ZJ, Duan YX (2020) Poly-adenine regulated DNA density on AuNPs to construct efficient DNA walker for microRNA-21 detection. Talanta 217:121056
He BS, Dong XZ (2021) Nb.BbvCI powered DNA walking machine-based Zr-MOFs-labeled electrochemical aptasensor using Pt@AuNRs/Fe-MOFs/PEI-rGO as electrode modification material for patulin detection. Chem Eng J 405:126642
Li W, Rong YC, Wang JY, Li TZ, Wang ZG (2019) MnO2 switch-bridged DNA walker for ultrasensitive sensing of cholinesterase activity and organophosphorus pesticides. Biosens Bioelectron 169:112605
Zhu X, Liu Y, Li P, Nie Z, Li J (2016) Applications of graphene and its derivatives in intracellular biosensing and bioimaging. Analyst 141(15):4541–4553
Ning Y, Zou L, Gao Q, Hu J, Lu FG (2018) Graphene oxide-based fluorometric determination of methicillin-resistant Staphylococcus aureus by using target-triggered chain reaction and deoxyribonuclease-assisted recycling. Microchim Acta 185(3):183
Jiang HY, Li FR, Li W, Lu XD, Ling K (2018) Multiplexed determination of intracellular messenger RNA by using a graphene oxide nanoprobe modified with target-recognizing fluorescent oligonucleotides. Microchim Acta 185(12):552
Wang W, Kong T, Zhang D, Zhang J, Cheng G (2015) Label-free microRNA detection based on fluorescence quenching of gold nanoparticles with a competitive hybridization. Anal Chem 91(18):11840–11847
Xiong HT, Zheng XW (2017) Electrochemiluminescence based determination of micro-RNA using target-guided assembly of gold nanoparticles on an electrode modified with Nafion, carbon nanotubes and polyvinylpyrrolidone. Microchim Acta 184(6):1781–1789
Wu H, Wang HY, Liu YL, Wu J, Zou P (2019) Fluorometric determination of microRNA by using target-triggered cascade signal amplification and DNA-templated silver nanoclusters. Microchim Acta 186:669
Liu L, Jiang S, Wang L, Zhang Z, Xie G (2015) Direct detection of microRNA-126 at a femtomolar level using a glassy carbon electrode modified with chitosan, graphene sheets, and a poly(amidoamine) dendrimer composite with gold and silver nanoclusters. Microchim Acta 182:77–84
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOC 4335 kb)
Rights and permissions
About this article
Cite this article
Ning, Y., Wang, X., Hu, J. et al. Graphene-based fluorometric determination of agrD gene transcription in methicillin-resistant Staphylococcus aureus using exonuclease III-aided target recycling and DNA walker cascade amplification. Microchim Acta 188, 269 (2021). https://doi.org/10.1007/s00604-021-04933-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04933-8