Abstract
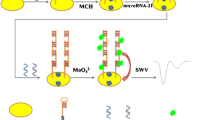
This article reports on a new square-wave anodic-stripping voltammetric method for sensitive determination of nucleic acids. It is based on a target-induced strand displacement reaction with blocker DNA (labeled with a CdS quantum dot) from a biotinylated hairpin DNA. First, a hairpin-blocker DNA duplex was immobilized on the surface of the well in a microtiter plate via biotin-streptavidin interaction. On addition of target DNA to the well, the CdS-labeled blocker DNA is displaced by target DNA from the hairpin-blocker duplex to form a new target-blocker DNA duplex. This is accompanied by the release of CdS-labeled blocker DNA. Next, cadmium ions are released from the Q-dots (by adding 1 M nitric acid) and then quantified by anodic stripping voltammetry using an in-situ prepared mercury film electrode. The voltammetric signal increases with the concentration of target DNA in the 5.0 pM to 1.0 nM concentration range, and the detection limit is as low as 1.2 pM. The assay has a good repeatability and displays an intermediate precision of down to 10 %.

The voltammetric nucleic acid assay is based on a target-induced strand displacement reaction between hairpin DNA and CdS quantum dot-labeled blocker DNA, and subsequent release and quantitation of Cd(II) ions.





Similar content being viewed by others
References
Khodakov D, Ellis A (2014) Recent developments in nucleic acid identification using solid-phase enzymatic assays. Microchim Acta 181:1633–1646
Chang K, Deng S, Chen M (2015) Novel biosensing methodologies for improving the detection of single nucleotide polymorphism. Biosens Bioelectron 66:297–307
Ikbal J, Lim G, Gao Z (2015) The hybridization chain reaction in the development of ultrasensitive nucleic acid assays. TrAC Anal Chem 64:86–99
Rodiger S, Liebsch C, Schmidt C, Lehmann W, Resch-Genger U, Schedler U, Schierack P (2014) Nucleic acid detection based on the use of microbeads: a review. Microchim Acta 181:1151–1168
Chen Y, Xiao L, Liu Y, Li X, Zhang J, Shu Y (2014) A lipase-based electrochemical biosensor for target DNA. Microchim Acta 181:615–621
Blackstock D, Chen W (2014) Halo-tag mediated self-labeling of fluorescent to molecular beacons for nucleic acid detection. Chem Commun 50:13735–13738
Gerasimova Y, Kolpashchikov D (2014) Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem Soc Rev 43:6405–6438
Pu F, Ren J, Ou X (2014) Nucleic acid and smart materials: advanced building blocks for logic systems. Adv Mater 26:5742–5757
Xu M, He Y, Gao Z, Chen G, Tang D (2015) Isothermal cycling and casecade signal amplification strategy for ultrasensitive colorimetric detection of nucleic acid. Microchim Acta 182:449–454
Shen J, Li Y, Gu H, Xia F, Zuo X (2014) Recent development of sandwich assay based on the nanobiotechnologies for proteins, nucleic acids, small molecules, and ions. Chem Rev 114:7631–7677
Wang F, Lu C, Willner I (2014) From casecade catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev 114:2881–2941
Zhu D, Zhang L, Ma W, Lu S, Xing X (2015) Detection of mircoRNA in clinical tumor samples by isothermal enzyme-free amplification and label-free graphene oxide-based SYBR Green I fluorescence platform. Biosens Bioelectron 65:152–158
Wong J, Yip S, Lee T (2014) Ultrasensitive and closed-tube colorimetric loop-mediated isothermal amplification assay using carboxyl-modified gold nanoparticles. Small 10:1495–1499
Lu W, Qin X, Luo X, Chang G, Sun X (2011) CdS quantum dots as a fluorescent sensing platform for nucleic acid detection. Microchim Acta 175:355–359
Wu Y, Xue P, Kang Y, Hui K (2013) Highly specific and ultrasensitive graphene-enhanced electrochemical detection of low-abundance tumor cells using silica nanoparticles coated with antibody-conjugated quantum dots. Anal Chem 85:3166–3173
Kokkinos C, Economou A, Petrou P, Kakabakos S (2013) Microfabricated tin-film electrodes for protein and DNA sensing based on stripping voltammetric detection of Cd(II) released from quantum dots labels. Anal Chem 85:10686–10691
Zhang B, Tang D, Goryacheva I, Niessner R, Knopp D (2013) Anodic-stripping voltammetric immunoassay for ultrasensitive detection of low-abundance proteins using quantum dot aggregated hollow microspheres. Chem Eur J 19:2496–2503
Lai W, Zhuang J, Tang J, Chen G, Tang D (2012) One-step electrochemical immunosensing for simultaneous detection of two biomarkers using thionine and ferrocene as distinguishable signal tags. Microchim Acta 178:357–365
Sun A, Jia F, Zhang Y, Wang X (2014) Gold nanocluster-encapsulated glucoamylase as a biolabel for sensitive detection of thrombin with glucometer readout. Microchim Acta. doi:10.1007/s00604-014-1440-1
Fu L, Tang D, Zhuang J, Lai W, Que X, Chen G (2013) Hybridization-induced isothermal cycling signal amplification for sensitive electronic detection of nucleic acid. Biosens Bioelectron 47:106–112
Zhu J, Ding Y, Liu X, Wang L, Jiang W (2014) Toehold-mediated strand displacement reaction triggered isothermal DNA amplification for highly sensitive and selective fluorescent detection of single-base mutation. Biosens Bioelectron 59:276–281
Qiu L, Shen Z, Wu Z, Shen G, Yu R (2015) Discovery of the unique self-assembly behavior of terminal suckers-contained dsDNA onto GNP and novel “light-up” colorimetric assay of nucleic acids. Biosens Bioelectron 64:292–299
Luo M, Li N, Liu Y, Chen C, Xiang X, Ji X, He Z (2014) Highly sensitive and multiple DNA biosensor based on isothermal strand-displacement polymerase reaction and functionalized magnetic microparticles. Biosens Bioelectron 55:318–323
Yu Z, Zaitonuna A, Lai R (2014) Effect of redox label tether length and flexibility on sensor performance of displacement-based electrochemical DNA sensor. Anal Chim Acta 812:176–183
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant no.: 21405128) and the Research (Initial) Fund for the Doctoral Program of Xinxiang University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sun, AL., Zhang, YF. & Wang, XN. Sensitive voltammetric determination of DNA via a target-induced strand-displacement reaction using quantum dot-labeled probe DNA. Microchim Acta 182, 1403–1410 (2015). https://doi.org/10.1007/s00604-015-1467-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-015-1467-y