Abstract
Lumbar spinal stenosis (LSS) is a lumbar disease with a high incidence in recent years. Accurate segmentation of the vertebral body, lamina and dural sac is a key step in the diagnosis of LSS. This study presents an lumbar spine magnetic resonance imaging image segmentation method based on deep learning. In addition, we define the quantitative evaluation methods of two clinical indicators (that is the anteroposterior diameter of the spinal canal and the cross-sectional area of the dural sac) to assist LSS diagnosis. To improve the segmentation performance, a dual-branch multi-scale attention module is embedded into the network. It contains multi-scale feature extraction based on three 3 × 3 convolution operators and vital information selection based on attention mechanism. In the experiment, we used lumbar datasets from the spine surgery department of Shengjing Hospital of China Medical University to evaluate the effect of the method embedded the dual-branch multi-scale attention module. Compared with other state-of-the-art methods, the average dice similarity coefficient was improved from 0.9008 to 0.9252 and the average surface distance was decreased from 6.40 to 2.71 mm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lumbar spinal stenosis (LSS) is a common degenerative disease in the elderly. With the aging of the population, the incidence rate has also increased significantly. At present, the clinical diagnosis of LSS is mainly based on the patient's symptoms, electrophysiology and imaging examination. Magnetic resonance imaging (MRI) and computed tomography (CT) are both considered as acceptable modalities in imaging [1]. MRI is the most frequently utilized imaging modality because it provides a detailed evaluation of the etiology and severity of spinal stenosis [2]. In the long-term practical work, clinicians found that some special imaging manifestations can objectively reflect stenosis, which is of great significance for the diagnosis of LSS. In addition to subjective evaluation of the spinal canal size, the commonly used quantitative measurement indicators are the anteroposterior diameter of the spinal canal, the area of the dural sac, and the distance between the ligamentum flavum at the level of the facet joint. Among them, the anteroposterior diameter of the spinal canal (SCAD) and the cross-sectional area of the dural sac (DSCA) are the most popular [3, 4]. Radiologists must diagnose each region of the spine through multiple slices of MRI images, and draw the defective areas by hand. However, it is difficult to separate the region of interest from MRI scans because this process requires simultaneous observation and exploration of large amounts of multimodal data to determine the lesion area. Therefore, it is necessary to establish a full-automatic auxiliary diagnostic system.
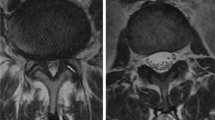
The following reasons make it difficult to correctly segment the spine (i.e. vertebral body, lamina, and dural sac) in MRI images. First, due to the low contrast of MRI images, the boundary between the spine and surrounding structures is often unclear, especially the boundary of the dural sac almost coincides with its adjacent background. As shown in Fig. 1 (row 1), the boundaries are difficult to distinguish even after careful contrast adjustment. Second, the internal distribution of the spinal structure (such as vertebral body and lamina) is uneven, as shown in Fig. 1 (row 2–3). Third, the shapes of spine have high variability and can change significantly across patients, as observed in the slices of three patients in Fig. 1 (row 1). Finally, simultaneous semantic segmentation of multiple spinal structures, are more difficult than individual tasks which will generate more complexity and indeterminacy.
Typical MRI images and their spine segmentation. The first row shows three different original MRI slices overlaid with their respective segmentation. Red denotes the lamina, yellow denotes the dural sac, and green denotes the vertebral body. The second and third rows show the original slice and gray histogram of the vertebral body and lamina respectively (color figure online)
To solve the problem mentioned above, researchers have proposed many algorithms for spine segmentation. For example, Naegel et al. [5] combined watershed and morphological method to segment spine. Ma et al. [6] manually determined the position of the spine in the image during the segmentation process, and then used the prior information such as the shape and the gradient introduced as constraints to achieve segmentation. Lim et al. [7] introduced the statistical shape of the spine as a priori information to initialize the level set function to improve the segmentation accuracy. Aslan et al. [8] constructed a new probability energy function that contains intensity, spatial interaction and shape information, and optimized this function to obtain the optimal segmentation. Although the above algorithms have achieved certain results, they have common problems: (1) The complexity of algorithms and the cumbersome segmentation process limit their application in the clinic; (2) The segmentation process requires manual intervention, and the performance depended on the design of manual feature; (3) The accuracy of each algorithm needs further improvement.
In recent years, with its powerful feature extraction and nonlinear modeling capabilities, convolutional neural networks (CNN) have been widely used in medical images such as CT, MRI, and ultrasound, and have achieved great success. Fan et al. [9] proposed a parallel reverse attention network for accurate polyp segmentation in colonoscopy images, which provided valuable information for polyp diagnosis and surgery. Imran et al. [10] presented a novel progressive adversarial semantic segmentation model, which can make improved segmentation predictions without requiring domain-specific data during training time, and they verified 8 public diabetic retinopathy and chest X-ray datasets. Chen et al. [11] devised a boundary-assisted region proposal network that achieves robust instance-level nucleus segmentation. Sara et al. [12] used Dense-Vnet to perform robust automatic whole brain extraction on skull striping common MRI sequences of brain tumor patients. Selvan et al. [13] treated the high opacity regions as missing data and presented a modified CNN-based image segmentation network that segmented lungs from such abnormal CXRs as part of a pipeline aimed at automated risk scoring of COVID-19 from CXRs. The latest achievements in spine segmentation research are: Amir et al. [14] showed that radiological gradings of spinal lumbar MRIs and pathological regions in the disc volumes can be achieved via a CNN framework. Han et al. [15] proposed a recurrent generative adversarial network for automated segmentation and classification of intervertebral discs, vertebrae, and neural foramen in MRIs in one shot. Ala et al. [16] used SegNet to aid clinicians in performing lumbar spinal stenosis detection through semantic segmentation and delineation of MRI scans of lumbar spine. Tang et al. [17] developed a dual densely connected U-shaped neural network to segment the spinal canal, dural sac and vertebral body in CT image to assist LSS diagnosis. To sum up, most of the lumbar spinal researchers based on deep learning more focus on the prediction performance and do not give specific clinical diagnostic criteria.
The contribution of this work is three-fold:
-
We constructed a new challenging data set with the experts of Spine Surgery of Shengjing Hospital to further study and evaluate the segmentation of spine T2-weighted MRI images;
-
We proposed a multi-scale attention U-shaped network (MANet) for semantic segmentation of the vertebral body, lamina, and dural sac. Specifically, the two convolutional layers between two down-sampling (or up-sampling) in the original U-Net are replaced by a convolutional layer and a dual-branch multi-scale attention module. The advantage of the module is that it adapts to different scale targets while retaining the key information of the image. This method can improve the efficiency of reading, reduce repetitive works, and eliminate inconsistencies like inter and intraobserver variance;
-
To assist in the diagnosis of LSS, we calculated the average cross-sectional area of the three anatomical structures, and we also accurately defined the DSCA for the first time. These clinical indicators can help doctors implement targeted treatment measures for patients.
The remainder of this paper is structured as follows: Sect. 2 covers the methodology part of this paper, where we introduce the architecture of MANet, explaining the details of the network training. In Sect. 3, visual comparison, qualitative and quantitative comparisons are conducted on the proposed method and state-of-the-art. Finally, the conclusion is described in Sect. 4.
2 Method
In this section, we will give a detailed description of the proposed framework for the segmentation of the vertebral body, lamina, and dural sac in MRI images. The whole architecture of the proposed framework is shown in Fig. 2
An illustration of the proposed framework for vertebral body, lamina, and dural sac segmentation in MRI images. The light grey cuboids represent feature maps, and the dark grey cuboids are copied feature maps from contracting path. The numbers below cuboid are the number of filter and the numbers above cuboid are the image resolution
2.1 Network architecture of proposed method
We design a deep fully convolutional network to segment vertebral body, lamina, and dural sac for the lumbar MRI images, which is a symmetric architecture like U-Net [18]. As illustrated in Fig. 2, it comprises of a contracting path to extract spatial features and an expanding path to restore image resolution. The contracting path contains four repeated stages, each stage involves a 3 × 3 convolution layers, a dual-branch multi-scale attention module and a max pooling layer with a pooling size of 2 × 2 and stride of 2. After each down-sampling, the filter number of the convolution layer are doubled. In the middle of network, a 3 × 3 convolution operation and a dual-branch multi-scale attention module connect the contracting path to the expanding path. Similarly, the expanding path also has four repeated stages. Contrary to the contracting path, the feature maps are up-sampled by using a 2 × 2 deconvolution operation first. The number of feature channels is reduced by half. After that, the corresponding results in the contracting path are copied and concatenated to the deconvolution results and follow this with a dual-branch multi-scale attention module. Finally, a 1 × 1 convolution operation is performed to generate the final segmentation map. All convolutional layers except for the last one use rectified linear unit (ReLU) activation function; the last convolution layer uses a softmax activation function.
2.2 Dual-branch multi-scale attention module
As shown in Fig. 3, the dual-branch multi-scale attention module consists of two branches. The upper branch is used for multi-scale feature extraction while the lower branch is responsible for screening key information. Given an intermediate feature map \(F \in \Re^{H \times W \times C}\) as input, the dual-branch multi-scale attention module infers a multi-scale feature map \(F_{M}\) and an attention map \(F_{A}\) in parallel. The final refined feature map \(F_{final}\) can be summarized as:
Dual-branch multi-scale attention Block. Dotted lines duplicate an intermediate feature map and feed it into the two branches separately. The upper branch is multi-scale feature extraction block, the colored boxes of which represent different scale feature maps and the lower branch is attention block
In this research, the vertebral body, lamina, and dural sac are the regions of interest, but in most cases, that are irregular and of different sizes. If a fixed-scale CNN is used, the range of the receptive field will be limited, which is not conducive to feature extraction. Multi-scale CNNs differ from fixed-scale CNNs by comprehensively using multiple scales to extract different information required for spine image segmentation. Currently, there are many cases of multi-scale CNN in natural images [19, 20], which can be summarized into three types: (1) methods using input images with different patch sizes and the same resolution; (2) methods using input images with the same region and different resolutions; (3) methods with different scale kernels. For the first two methods, the input image and the corresponding ground truth have different resolutions, they cannot be directly put into a CNN, and different input data needs to be prepared. The third method only needs to train multi-scale CNN with different kernel sizes for segmentation. It also can avoid the problem of fixed-scale CNN's limitation on the receptive field, and can extract features at multiple scales, which is in favor of improve the accuracy of image segmentation. Therefore, we followed the idea of the third method that is to concatenate three 3 × 3 convolution operations [21] to construct a multi-scale feature extraction structure equivalent to extracting features from 3 × 3, 5 × 5 and 7 × 7 convolution operation simultaneously.
The feature maps will lose a great deal of details and edge information after a series of convolution and pooling, nevertheless, the attention mechanism [22] will compensate for this information to a certain extent. The attention mechanism is an image processing technology that learns from human vision. Therefore, we first briefly introduce the selective attention mechanism of human vision. Human vision quickly scans the global image to obtain the target area that needs to be focused, and then invests more attention resources in this area to get more detailed information about the target, while suppressing other useless information. The attention mechanism in deep learning is similar to the selective attention mechanism of human vision in essence. The core goal is to select information that is more critical to the current task from a huge amount of information.
As shown in the lower branch of Fig. 3, the attention block merge the channel domain and spatial domain. The attention block [23] computes a channel attention map \(M_{C} \in 1 \times 1 \times C\) and a spatial attention map \(M_{S} \in H \times W \times 1\) sequentially. \(M_{C}\) gives the meaningful semantics by exploiting the inter-channel relationship of features. To compute \(M_{C}\), the average pooling and max pooling operations are used to aggregate the spatial information of the feature map, thereby generating two different spatial context descriptors, which represent average pooled features \(F_{C}^{a}\) and max pooled features \(F_{C}^{m}\) respectively. After that \(F_{C}^{a}\) and \(F_{C}^{m}\) are forwarded to a multi-layer perceptron (MLP) with one hidden layer. After the MLP is applied, we merge the two output feature vectors using element-wise summation and then perform a sigmoid function. The channel attention is computed as:
where \(\sigma\) denotes the sigmoid function and \(w_{MLP}\) is the MLP weights shared for both inputs.
Spatial attention block utilizes the inter-spatial relationship to generate \(M_{S}\), which is different from channel attention block.\(M_{S}\) focuses on the position of the features, which is complementary to the channel attention. To compute \(M_{S}\), we apply average pooling and max pooling operations along the channel axis, and concatenate them to generate an effective feature descriptor. After which a convolution operation with the filter size of 7 × 7 and a sigmoid function are used. The calculation process is as follows:
where \(\sigma\) denotes the sigmoid function, \(F_{C}\) is refined feature map computed by channel attention block, \(f\left( \cdot \right)\) is a convolution operation with the filter size of 7 × 7 and \(F_{S}^{a}\), \(F_{S}^{m}\) represent average pooled features and max pooled features respectively.
The entire attention block can be expressed as:
where \(F_{A}\) is the final refined output and \(\otimes\) denotes element-wise multiplication.
2.3 Backbone network
Neural network training requires large amounts of datasets and expensive calculation costs, such as AlexNet [24], VGG [25] and other models easily have hundreds of millions of parameters. For specific problems, due to the limitation of training cost, the ideal neural network architecture cannot be fully realized. Accordingly, we often need to use some optimization techniques to compress the size of the model. In this paper, we reduce the parameters of the original U-Net model (which has about 28 million parameters) by adjusting the depth of model or the channel of convolutional layer. We set the number of feature channels of the first convolution layer from 64 to 16. Furthermore, after each convolution layer, batch normalization (BN) is applied to reduce the total parameters of the model to 4.6 million. In detail, we extract the multi-scale pixel-level attention feature maps with the encoder, and the size of the output feature map is 1/16th of the input image. Combined the high-level features and the low-level features via skip-connections to generate the final predicted map.
2.4 Training
We randomly divided the dataset into three parts, namely 70% for training, 20% for validation, and 10% for testing. In practice, 70% of the images are input to the MANet for training, 20% of the images are used for hyperparameter optimization and preventing of overfitting, and 10% of the images are used to evaluate the performance of the neural network. The MANet is trained in an end-to-end manner when optimizing all parameters in the network. The task of semantic segmentation is to predict whether a pixel represents a point of interest, or just a part of background. Thus, the problem in this paper can be reduced to a pixel-wise multi-class segmentation problem. In summary, we can choose a loss function to directly optimize the evaluation criteria. Dice loss is a good option, but for a typical lumbar spine MRI image most pixels are the background, and the areas of the vertebral body, lamina, and dural sac are extremely small and have different proportions. Dice loss is very detrimental to the small target, because in the case of only the foreground and the background, once the small target has some pixel prediction errors, it will cause a large change in loss, resulting in a sharp gradient change and unstable training. To overcome this problem, we introduced a hybrid loss function, which is composed of the generalized dice loss [26] and the cross-entropy loss. It assigns different weights to the dice loss of each class and combines the cross entropy of each image. The hybrid loss function compensates for the imbalance between the large and the small target and promotes the neural network to learn the features of the small tissue such as the dural sac. Specifically, this loss function can be expressed as:
where \(L_{hy}\) is hybrid loss, \(L_{GD}\) is generalized dice loss and \(L_{CE}\) is cross-entropy loss. m represents the class, i.e., vertebral body (m = 1), lamina (m = 2), dural sac (m = 3) and background (m = 4), \(w_{j}\) donates the class-balancing weight, N is pixel number of the image,\({\text{y}}_{ij}\) is the ground truth of the pixel i belonging to class j, and \({\hat{\text{y}}}_{ij}\) is the corresponding predicted probability value of \({\text{y}}_{ij}\).
The code of MANet is implemented by Python 2.7 and Keras 2.2.4, and our model was trained and tested on a Nvidia GeForce GTX TITAN X GPU, developed on a 64-bit ubuntu 14.04 platform with Intel Core i7-5930K CPU with 64 GB RAM. Due to GPU memory constraints, our model is trained with a mini-batch size of 4. The optimizer that we adopt is stochastic gradient descent (SGD) [27] with momentum coefficient is set to 0.9 and the initial learning rate (lr) is set to 0.001. The learning rate varies with the training epochs, when training epochs is 200, the decay argument is specified, decay = 0.1/epochs, and the learning rate of each training is decreased to lr = r/(1 + decay × epoch). We randomly initialize parameter weights according to the Xavier scheme.
3 Experiments and results
In this section, we introduce the dataset provided by the spine surgery department of Shengjing Hospital of China Medical University and performance evaluation metrics of this article, after which we discuss the effectiveness of the MANet with U-Net as the baseline, then we compare the MANet with the state-of-the-art methods. At last we calculate the cross-sectional area (CSA) of vertebral body, lamina and the dural sac and the distance of the anteroposterior diameter of the spinal canal.
3.1 Data sources
Although previous studies have attempted to simultaneously segment the vertebral body, lamina, and dural sac from the MRI image, there is no public dataset. Therefore, we built a new pixel-level label dataset with the spine surgery department of Shengjing Hospital of China Medical University. We collected axial T2-weighted lumbar MRI images of L3-4, L4-5 and L5-S1 discs of young male patients aged 18 to 35 in the same hospital. In order to ensure the consistency of the label, we performed brightness, contrast adjustment and normalization processing, and the size of all images was unified to 512 × 512. Four spine surgeons and two imaging surgeons used Photoshop graphics software to label the vertebral body, lamina and dural sac in the image manually, which were double-checked by one spine surgery specialist with more than 30 years of experience and two spine surgeons with more than 10 years of experience. Ultimately, we retained 1080 images of 120 patients with precise and consistent pixel-level labels. We randomly divided the dataset into three parts, 70% of which is for training, 20% for validation, and 10% for testing.
3.2 Metrics
In the semantic segmentation of this paper, the region of vertebral body, lamina and dural sac only comprise a small part of the entire image. Therefore, metrics such as precision and recall are inadequate and often lead to a false sense of superiority, inflated by the perfect background detection. Hence, we evaluate the performance of model with two widely used evaluation criterion of medical image segmentation: Dice similarity coefficient (DSC) and average surface distance (ASD). DSC is a function to evaluate the similarity, which is used to calculate the similarity or overlap of two samples:
where \(V_{{{\text{gt}}}}\) and \(V_{{{\text{seg}}}}\) denote the pixel sets of the manually labeled ground truth and automatically segmented spinal structure, respectively. The value range of DSC is [0, 1], the higher the DSC, the higher the similarity between the segmentation result and ground truth, that is, the better the segmentation performance.
ASD evaluates the symmetrical mean distance between two samples:
where \(d\left( \cdot \right)\) is the Euclidean Distance. The higher the value of ASD, the lower the matching degree of the two samples. This metric can also be called average symmetric surface distance (ASSD).
3.3 Comparison with U-Net
As shown in Tables 1 and 2 (from 5th row to 6th row), the dual-branch multi-scale attention module endows MANet a superior performance for the segmentation of vertebral body, lamina, and dural sac. As a baseline, U-Net on average achieves 0.9008 DSC and 6.40 mm ASD. After only preserving the upper branch (Multi-scale branch), DSC is increased by 1.51% and ASD is decreased by 2.38 mm. This demonstrates the upper branch can obtain semantic representation of different scale targets. Then, after preserving the lower branch (attention branch), DSC and ASD are 0.9152 and 3.68 mm, an increase of 1.44% and a decrease of 2.72 mm respectively, which proves the lower branch can effectively correct the errors of semantic segmentation by suppressing background information. Finally, after preserving the whole dual-branch multi-scale attention module (MANet), the DSC and ASD are greatly changed, which demonstrates the effectiveness of the dual-branch multi-scale attention module.
Then we observed the performance of the model with 200 epochs. In Fig. 4, the performance of the validation data on each epoch is shown. We give the average accuracy index and average loss value of each epoch. It is worth noting that for all cases, our proposed model can quickly converge. This can be attributed to dual-branch multi-scale attention feature extraction. Another notable finding in the experiment is that, except for some minor change, the deviation of the MANet is much smaller, that shows the reliability and robustness of the proposed model. Besides, these results show that, compared with the classic U-Net, the proposed network may obtain better results with fewer training epochs.
U-Net has achieved satisfactory results in medical image segmentation, but with in-depth study of some images, especially those with less clear boundaries, U-Net seems to be struggling a bit. We marked the edges of MANet and U-Net for comparison. As shown in Fig. 5a, b and e, f, when we segment the vertebral body and dural sac, U-Net does not accurately locate the edge, however, the performance of MANet is nearly perfect. When segmenting the lamina, neither method performs well, but our method is still slightly better, which is shown in Fig. 5c, d.
Segmenting spine structure with clearly visiable boundary. The first row is original spine MRI images. The second row is the enlarged segmentation result with annotations. Red represents ground truth, blue represents segmentation results from MANet and green represents segmentation from U-Net (color figure online)
The segmentation task of this paper is to cluster the homologous pixels in the spine MRI image. However, in real medical images, distinguishing the region of interest from the background is challenging, so we will face two segmentation extremes: (1) The result is not a continuous segmentation region, but a collection of fractured segmentation regions. (2) Due to texture and disturbance, sometimes the background looks so similar to the foreground, and some results that do not belong to the target are obtained. These two situations lead to information loss and classification errors, respectively. Figure 6 shows the third part of the experiment, we compare the performance of the algorithms in extreme cases. When segmenting the lamina or vertebral body, if there is a slight change in the foreground object, U-Net cannot segment the target into a continuous region (shown in Fig. 6a, b). It predicts a set of scattered regions, confuses the target as background, and thus loses some valuable information. On the other hand, for images with uneven background, the U-Net model seems to make some false predictions shown in Fig. 6d–f. In addition, in some extremely unfavorable situations, the U-Net model cannot make any predictions at all because the difference between the target and the background is too subtle. Although the segmentation of MANet is not perfect in this challenging situation, its performance is far superior to the classic U-Net model.
3.4 Comparison with other state-of-the-art methods
To further verify the performance of the algorithms, we tested the proposed method on the collected dataset and compared the performance with four state-of-the-art image segmentation methods, i.e. FCN [28], U-Net, Res2net [29] and Deeplabv3 + [30]. Both visual comparison and quantitative comparison show that our method is superior to these advanced methods. The comparison results in Tables 1, 2 and Fig. 7 show the advantages of the MANet algorithm. Compared with the existing segmentation network, MANet is significantly better than the FCN and U-Net network by 2.15% and 2.44% average DSC and 2.7 mm and 3.69 mm average ASD. MANet outperforms the Deeplabv3 + network by 1.41% average DSC and 1.44 mm average ASD. Also, MANet beat Res2net network by 1.08% average DSC and 0.5 mm average ASD. Therefore, MANet has strong predictive performance and application ability in the computer-aided diagnosis of LSS.
Visual comparison between MANet and four state-of-the-art methods. The images in the rightmost column are the ground truth of each row, where the green regions indicate vertebral body, yellow regions indicate the dural sac, blue regions indicate the lamina, and dark regions indicate the background. From b to f are the results of FCN, U-Net, Res2net, DeeplabV3 + and the proposed MANet, respectively. Our results are the closest to the ground truth (g) (color figure online)
Overall, the proposed MANet model can successfully segment the spinal structure in most cases, but its segmentation performance has certain limitations. For example, some patients have suffered from LSS for a long time, the spine structure has undergone severe deformation, the vertebral body or lamina and the adjacent soft tissues are connected or the dural sac is seriously compressed. In the above cases, the MANet model cannot accurately segment the spinal structure, especially the vertebral body. Figure 8 shows some examples of poor MANet segmentation on these difficult slices.
Examples of poor MANet segmentation on difficult slices. a–b There is adhesion between the vertebral body and the paraspinal muscles, which makes their edges ambiguous or imperceptible. c The dural sac compressed by the vertebral body and lamina is impossible to segment accurately. d Severely deformed and blurred lamina is difficult to separate from background. The red dotted line represents the boundary of the anatomical structure, and the red rectangular box is the region of interest (color figure online)
3.5 Measurement of clinical indicators
In order to assist doctors in diagnosing LSS, in the last part of the experiment, we measured the cross-sectional area of the vertebral body, lamina, and dural sac as well as the anteroposterior diameter distance of the spinal canal with 108 test images. After training the proposed network, we return the probability value of each pixel as the output result. By binarizing all pixel values, a binary image with target pixel of 1 and background pixel of 0 can be obtained. In MRI slice, we calculate the number of pixels with the gray value of 1 in the binary image to get the cross-sectional area of a target. Under the premise that the pixel resolution is known, the physical area of the region of interest can be obtained. The whole process is as follows:
where Input is the input image, and Output is the output image whose value is the probability. B is the binarized output image, Num is the pixel area of target, u is the physical length of the pixel and A is the physical area of target.
Figure 9 shows several cases of the anteroposterior diameter of the spinal canal. When calculating the anteroposterior diameter of the spinal canal, we approximate this distance to the distance from the center of the bottom of the vertebral body to the most concave point on the upper edges of the lamina. In the first step, we need to calculate the center of the bottom of the vertebral body. In this process, we first find the center of the vertebral body, and regard the horizontal coordinate of the center as the horizontal coordinate of the bottom center of the vertebral body. Then we compute the vertical coordinate corresponding to this horizontal coordinate on the curve of the vertebral body contour and regard the position with the smaller absolute value as the vertical coordinate of the bottom center of the vertebral body. This coordinate can be expressed as follows:
where \(C_{VB}\) is outline of vertebral body, \(\left( {Cen_{x} ,Cen_{y} } \right)\) is the center of vertebral body and \(\left( {Cen_{x} ,Bottom_{y} } \right)\) is the bottom center of vertebral body. getCenLocation function represents to extract the center of a contour, and findCoo_y function represents to find a vertical coordinate in the contour.
After which, we find the most concave point on the upper edge of the lamina. Eventually, we get the anteroposterior diameter of the spinal canal by calculating the geometrical distance of the two point. The specific process is as follows:
where \(C_{L}\) is outline of the lamina, \(E_{upper}\) is the upper edge of the lamina, \(\left( {Conca_{x} ,Conca_{y} } \right)\) is the most concave point on the upper edge of the lamina, and D is the anteroposterior diameter of the spinal canal. getE_upper function represents to extract the upper edge of a contour and getCoo_cancave function represents to get coordinate of the most concave point of a curve.
To validate the consistency between the calculated clinical indicators of the predicted results and the clinical indicators obtained from manually annotation, we used the linear regression equations to show the correlation of several sets of clinical indicators. The linear regression equation is calculated as follow:
where X is the predicted results, and Y is ground truth.\(a_{1}\) is the constant term of the overall regression equation, which is the intercept of the overall regression line on the Y axis; \(a_{0}\) is the overall regression coefficient, and is also the slope of the overall regression line.
where \(\overline{X}\) is the mean of predicted results, and \(\overline{Y}\) is the mean of ground truth.\(X_{i}\) and \(Y_{i}\) are the corresponding samples in X and Y respectively. The closer the regression curve is to \(Y = X\), the closer the predicted result is to the ground truth.
Figure 10 shows the linear regression curve of the cross-sectional area of the vertebral body, lamina, dural sac, and the anteroposterior distance of the spinal canal. The regression coefficients are 1.0186, 0.872, 1.0275, and 1.0435, respectively, which shows that the prediction results are highly consistent with manual annotations, which also proves that the network we proposed is effective and can be applied to the clinic.
a–c The linear regression curve of the cross-sectional area of the vertebral body, lamina, dural sac. d The linear regression curve of the distance of the anteroposterior distance of the spinal canal. The red curve is regression curve and the horizontal and vertical coordinates of blue dot represent ground truth and predicted result respectively (color figure online)
4 Conclusion
Precise segmentation of the vertebral body, lamina and dural sac is a key step in the diagnosis of LSS. This paper proposed a new spine MRI image segmentation method and calculated the CSA of the spinal structure and the DASC. Compared with several commonly used medical image segmentation methods, the proposed method has achieved better segmentation results. The method was tested on real spine MRI data and evaluated through similarity metrics such as dice similarity coefficient and average surface distance. The results of these similarity metrics were 92.52% and 2.71 mm respectively. These results prove the effectiveness of our method.
In short, the main contribution of this paper is to propose a dual-branch multi-scale attention module which can extract different information required for spine image segmentation and select key information in feature maps. Quantitative and qualitative experimental results show that our method improves segmentation accuracy and corrects segmentation error. The results also show higher overlap and lower distance between the automatic segmentation and manual annotation.
In the future, we intend to use broad training data to evaluate our method, such as low-contrast data, elderly data, and lumbar disease patients’ data to improve performance. The other direction is to speed up the calculation time of the training stage through optimization. Although our model has performed a channel pruning, the training parameters and computational complexity have been reduced to a certain extent, but it still takes more than 20 h to train 200 epochs with a GPU. In this regard, reducing the calculation time of the training stage will be the future work.
Abbreviations
- LSS:
-
Lumbar spinal stenosis
- MRI:
-
Magnetic resonance imaging
- CT:
-
Computed tomography
- SCAD:
-
Anteroposterior diameter of the spinal canal
- CSA:
-
Cross-sectional area
- CNN:
-
Convolutional neural network
- DSCA:
-
Cross-sectional area of the dural sac
- ReLU:
-
Rectified linear unit
- MLP:
-
Multi-layer perceptron
- BN:
-
Batch normalization
- SGD:
-
Stochastic gradient descent
- GT:
-
Ground truth
References
Graaf ID, Prak A, Bierma-Zeinstra S et al (2006) Diagnosis of lumbar spinal stenosis: a systematic review of the accuracy of diagnostic tests. Spine 31(10):1168–1176
Cheng F, You J, Rampersaud YR (2010) Relationship between spinal magnetic resonance imaging findings and candidacy for spinal surgery. Can Fam Physician 56(9):e323
Schönström M, Lindahl S, Jan W et al (1989) Dynamic changes in the dimensions of the lumbar spinal canal: an experimental study in vitro. J Orthop Res 7(1):115–121
Ogikubo O, Forsberg L, Hansson T (2007) The relationship between the cross-sectional area of the cauda equina and the preoperative symptoms in central lumbar spinal stenosis. Spine 32(13):1423
Naegel B (2007) Using mathematical morphology for the anatomical labeling of vertebrae from 3D CT-scan images. Comput Med Imaging Graph 31(3):141–156
Ma J, Lu L (2013) Hierarchical segmentation and identification of thoracic vertebra using learning-based edge detection and coarse-to-fine deformable model. Comput Vis Image Underst 117(9):1072–1083
Lim PH, Bagci U, Bai L (2013) Introducing Willmore flow into level set segmentation of spinal vertebrae. IEEE Trans Biomed Eng 60(1):115–122
Melih A, Ahmed S, Asem M et al (2015) Model-based segmentation, reconstruction and analysis of the vertebral body from spinal CT. Comput Vis Biomech 18:381–437
Fan DP, Ji GP, Zhou T et al (2020) PraNet: parallel reverse attention network for polyp segmentation. arXiv:2006.11392
Imran AAZ, Terzopoulos D (2020) Progressive adversarial semantic segmentation. arXiv:2005.04311
Chen S, Ding C, Tao D (2020) Boundary-assisted region proposal networks for nucleus segmentation. arXiv
Ranjbar S, Singleton KW, Curtin L, Rickertsen CR, Paulson LE, Hu LS, Mitchell JR, Swanson KR (2020) Robust automatic whole brain extraction on magnetic resonance imaging of brain tumor patients using dense-Vnet. ArXiv abs/2006.02627
Selvan R, Dam EB, Rischel S et al (2020) Lung segmentation from chest X-rays using variational data imputation. arXiv:2005.10052
Jamaludin A, Kadir T, Zisserman A (2017) SpineNet: automated classification and evidence visualization in spinal MRIs. Med Image Anal 41:63
Han Z, Wei B, Mercado A, Leung S, Li S (2018) Spine-GAN: semantic segmentation of multiple spinal structures. Med Image Anal 50:23–35
Al-Kafri AS, Sudirman S, Hussain AJ, Al-Jumeily D et al (2019) Boundary delineation of MRI images for lumbar spinal stenosis detection through semantic segmentation using deep neural networks. IEEE Access 7:43487–43501
Tang H, Xiaobing P, Shilong H, Xin L et al (2020) Automatic lumbar spinal CT image segmentation with a dual densely connected U-Net. IEEE Access. arXiv:1910.09198
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. Comput Sci. arXiv:1505.04597v1
Ji Y, Zhang H, Jonathan WQM (2018) Salient object detection via multi-scale attention CNN. Neurocomputing 322:130–140
Dong L, Zhang H, Ji Y et al (2020) Crowd counting by using multi-level density-based spatial information: a multi-scale CNN framework-ScienceDirect. Inform Sci 528:79–91
Lou A, Guan S, Loew M (2020) DC-UNet: rethinking the U-Net architecture with dual channel efficient CNN for medical images segmentation. arXiv:2006.00414
Xu K, Ba J, Kiros R et al (2015) Show, attend and tell: neural image caption generation with visual attention. Comput Sci 2048–2057. arXiv:1502.03044v3
Woo S, Park J, Lee JY et al (2018) CBAM: convolutional block attention module. arXiv:1807.06521
Krizhevsky A, Sutskever I et al (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. Comput Sci. arXiv:1409.1556v4
Sudre CH, Li W, Vercauteren T et al (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv:1707.03237
Sutskever I (2013) On the importance of initialization and momentum in deep learning. In: International conference on machine learning, pp 1139–1147
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651
Gao S, Cheng M, Zhao K, Zhang X, Yang M et al (2019) Res2Net: a new multi-scale backbone architecture. IEEE Trans Pattern Anal Mach Intell 1–1. arXiv:1904.01169v1
Liang CC, Yukun Z, George P, Florian S et al (2018) Encoder–decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp 8–14
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, H., Luo, H., Huan, W. et al. Automatic lumbar spinal MRI image segmentation with a multi-scale attention network. Neural Comput & Applic 33, 11589–11602 (2021). https://doi.org/10.1007/s00521-021-05856-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-05856-4