Abstract
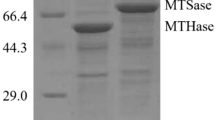
Maltooligosyl trehalose trehalohydrolase (MTHase, EC 3.2.1.141) catalyzes the release of trehalose, a novel food ingredient, by splitting the α-1,4-glucosidic linkage adjacent to the α-1,1-glucosidic linkage of maltooligosyl trehalose. However, the high-yield preparation of recombinant MTHase has not yet been reported. In this study, a codon-optimized synthetic gene encoding Sulfolobus acidocaldarius MTHase was expressed in Escherichia coli. In initial expression experiments conducted using pET-24a (+) and E. coli BL21 (DE3), the MTHase activity was 10.4 U/mL and a large amount of the expression product formed inclusion bodies. The familiar strategies, including addition of additives, co-expression with molecular chaperones, and expression with a fusion partner, failed to enhance soluble MTHase expression. Considering the intermolecular disulfide bond of MTHase, expression was investigated using a system comprising plasmid pET-32a (+) and host E. coli Origami (DE3), which is conducive to cytoplasmic disulfide bond formation. The MTHase activity increased to 55.0 U/mL, a 5.3-fold increase. Optimization of the induction conditions in a 3-L fermentor showed that when the lactose was fed at 0.2 g/L/h beginning at an OD600 of 40 and the induction temperature was maintained at 30 °C, the MTHase activity reached a maximum of 204.6 U/mL. This is the first report describing a systematic effort to obtain high-efficiency MTHase production. The high yield obtained using this process provides the basis for the industrial-scale production of trehalose. This report is also expected to be valuable in the production of other enzymes containing disulfide bonds.





Similar content being viewed by others
References
Fang TY, Tseng WC, Shih TY, Wang MY (2008) Identification of the essential catalytic residues and selectivity-related residues of maltooligosyltrehalose trehalohydrolase from the thermophilic archaeon Sulfolobus solfataricus ATCC 35092. J Agric Food Chem 56:5628–5633
Kato M (1999) Trehalose production with a new enzymatic system from Sulfolobus solfataricus KM1. J Mol Catal B Enzym 6:223–233
Richards AB, Krakowka S, Dexter LB, Schmid H, Wolterbeek APM, Waalkens-Berendsen DH, Shigoyuki A, Kurimoto M (2002) Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem Toxicol 40:871–898
Ohtake S, Wang YJ (2011) Trehalose: current use and future applications. J Pharm Sci US 100:2020–2053
Sebaaly C, Greige-Gerges H, Stainmesse S, Fessi H, Charcosset C (2016) Effect of composition, hydrogenation of phospholipids and lyophilization on the characteristics of eugenol-loaded liposomes prepared by ethanol injection method. Food Biosci 15:1–10
Mukai K, Tabuchi A, Nakada T, Shibuya T, Chaen H, Fukuda S, Kurimoto M, Tsujisaka Y (1997) Production of trehalose from starch by thermostable enzymes from Sulfolobus acidocaldarius. Starch Starke 49:26–30
Kim YH, Kwon TK, Park S, Seo HS, Cheong JJ, Kim CH, Kim JK, Lee JS, Choi YD (2000) Trehalose synthesis by sequential reactions of recombinant maltooligosyltrehalose synthase and maltooligosyltrehalose trehalohydrolase from Brevibacterium helvolum. Appl Environ Microbiol 66:4620–4624
Yamamoto T, Maruta K, Watanabe H, Yamashita H, Kubota M, Fukuda S, Kurimoto M (2001) Trehalose-producing operon treYZ from Arthrobacter ramosus S34. Biosci Biotechnol Biochem 65:1419–1423
Nakada T, Maruta K, Mitsuzumi H, Kubota M, Chaen H, Sugimoto T, Kurimoto M, Tsujisaka Y (1995) Purification and characterization of a novel enzyme, maltooligosyl trehalose trehalohydrolase, from Arthrobacter sp Q36. Biosci Biotechnol Biochem 59:2215–2218
Nakada T, Ikegami S, Chaen H, Kubota M, Fukuda S, Sugimoto T, Kurimoto M, Tsujisaka Y (1996) Purification and characterization of thermostable maltooligosyl trehalose trehalohydrolase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biosci Biotechnol Biochem 60:267–270
Demain AL, Vaishnav P (2009) Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv 27:297–306
Schiraldi C, Di Lernia I, De Rosa M (2002) Trehalose production: exploiting novel approaches. Trends Biotechnol 20:420–425
de Pascale D, Sasso MP, Di Lernia I, Di Lazzaro A, Furia A, Farina MC, Rossi M, De Rosa M (2001) Recombinant thermophilic enzymes for trehalose and trehalosyl dextrins production. J Mol Catal B Enzym 11:777–786
Seo JS, An JH, Baik MY, Park CS, Cheong JJ, Moon TW, Park KH, Choi YD, Kim CH (2007) Molecular cloning and characterization of trehalose biosynthesis genes from hyperthermophilic archaebacterium Metallosphaera hakonesis. J Microbiol Biotechnol 17:123–129
Chang M, Qiao Y, Ding HB (2011) Cloning and expression of maltooligosyltrehalose trehalohydrolase gene from Corynebacterium glutamicum. J Agri Sci Technol 13:47–52
Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4:1
Barroso JF, Elholm M, Flatmark T (2003) Tight binding of deoxyribonucleotide triphosphates to human thymidine kinase 2 expressed in Escherichia coli. Purification and partial characterization of its dimeric and tetrameric forms. Biochemistry 42:15158–15169
Okamoto-Kainuma A, Yan W, Fukaya M, Tukamoto Y, Ishikawa M, Koizumi Y (2004) Cloning and characterization of the dnaKJ operon in Acetobacter aceti. J Biosci Bioeng 97:339–342
Blackwell JR, Horgan R (1991) A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett 295:10–12
Kagawa N, Cao QW (2001) Osmotic stress induced by carbohydrates enhances expression of foreign proteins in Escherichia coli. Arch Biochem Biophys 393:290–296
Duan X, Chen J, Wu J (2013) Optimization of pullulanase production in Escherichia coli by regulation of process conditions and supplement with natural osmolytes. Bioresour Technol 146:379–385
Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR (2004) SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genom 5:75–86
Peroutka Iii RJ, Orcutt SJ, Strickler JE, Butt TR (2011) SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes. Methods Mol Biol 705:15–30
Smith DB, Johnson KS (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31–40
Lavallie ER, Diblasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the Escherichia coli cytoplasm. Biotechnology 11:187–193
Han C, Su LQ, Hong RY, Wu SX, Wu J (2017) A comparative study of maltooligosyltrehalose synthase from Sulfolobus acidocaldarius expressed in Pichia pastoris and Escherichia coli. Process Biochem 60:35–41
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal chem 31:426–428
Bessette PH, Aslund F, Beckwith J, Georgiou G (1999) Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA 96:13703–13708
de Marco A (2009) Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact 8:26
Cheng J, Wu D, Chen S, Chen J, Wu J (2011) High-level extracellular production of alpha-cyclodextrin glycosyltransferase with recombinant Escherichia coli BL21 (DE3). J Agric Food Chem 59:3797–3802
Su LQ, Hong RY, Wu J (2015) Enhanced extracellular expression of gene-optimized Thermobifida fusca cutinase in Escherichia coli by optimization of induction strategy. Process Biochem 50:1039–1046
Su LQ, Huang Y, Wu J (2015) Enhanced production of recombinant Escherichia coli glutamate decarboxylase through optimization of induction strategy and addition of pyridoxine. Bioresource Technol 198:63–69
Donovan RS, Robinson CW, Glick BR (1996) Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol 16:145–154
Su LQ, Ma Y, Wu J (2015) Extracellular expression of natural cytosolic arginine deiminase from Pseudomonas putida and its application in the production of L-citrulline. Bioresour Technol 196:176–183
Kilikian BV, Suarez ID, Liria CW, Gombert AK (2000) Process strategies to improve heterologous protein production in Escherichia coli under lactose or IPTG induction. Process Biochem 35:1019–1025
Lebendiker M, Danieli T (2014) Production of prone-to-aggregate proteins. FEBS Lett 588:236–246
Acknowledgements
This work received financial support from the National Natural Science Foundation of China (31771916, 31501419), the Natural Science Foundation of Jiangsu Province (BK20180082), the National Science Fund for Distinguished Young Scholars (31425020), the National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-03), and the 111 Project (No. 111-2-06).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
The codon-optimized synthetic gene (treZ) sequence (TIFF 4432 KB)
Fig. S2
The construction diagram of recombinant plasmid pET24a-treZ (TIFF 3362 KB)
Fig. S3
The construction diagram of recombinant plasmid pET24a-sumo-treZ (TIFF 4512 KB)
Fig. S4
Effects of different additives on cell growth and MTHase production. ■ OD600, □ MTHase activity (TIFF 35 KB)
Fig. S5
SDS-PAGE analysis of recombinant MTHase in the 3-L fermentor. M, molecular mass standard proteins; 1–8, the MTHase samples corresponding with that in Table 1 (TIF 6446 KB)
Rights and permissions
About this article
Cite this article
Su, L., Wu, S., Feng, J. et al. High-efficiency expression of Sulfolobus acidocaldarius maltooligosyl trehalose trehalohydrolase in Escherichia coli through host strain and induction strategy optimization. Bioprocess Biosyst Eng 42, 345–354 (2019). https://doi.org/10.1007/s00449-018-2039-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-018-2039-4