Abstract
Genome editing through the alteration of nucleotide sequence has already revolutionized the field of site-directed mutagenesis for a decade. However, research in terms of precision and efficacy in targeting the loci and reduction in off-target mutation has always been a priority when DNA is involved. Therefore, recent research interest lies in utilizing the same precision technology but results in non-transgenic. In this review article, different technological advancements have been explained, which may provide a holistic concept of and need for transgene-free genome editing. The advantage and lacunas of each technology have been critically discussed to deliver a transparent view to the readers. A systematic analysis and evaluation of published research articles implied that researchers across the globe are putting continuous efforts in this direction to eliminate the hindrance of transgenic regulation. Nevertheless, this approach has severe implications legitimate for mitigating the conflict of acceptance, reliability, and generosity of gene-editing technology and sustainably retorting to the expanding global population feeding challenges.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated) system has been performing as the most promising editing tool in the era of genomics with respect to the identification of gene functionality and crop improvement. Ideally, CRISPR refers to tandem repeats flanked by non-repetitive nucleotide stretches that were first observed downstream of the iap gene of Escherichia coli (Ishino et al. 1987). These non-repetitive sequences were identical to foreign DNA sequences derived from phages and plasmids, which serve as an adaptive immune system in bacteria and archaea.
To date, most accepted programmable nucleases utilize the CRISPR/Cas system, and the target specificity is governed by a so-called short CRISPR-RNA (crRNA). It is encoded in the CRISPR-locus. Another component, Protospacer-adjacent motif (PAM), generally 5’ NGG, is required for the correct recognition of the target site (Jinek et al. 2012). The trans-activating short CRISPR-RNA (tracrRNA) fuses with crRNA (together called single-guide RNA, sgRNA) to form a stable complex with Cas9, which eventually edits the targeted region of nucleic acid.
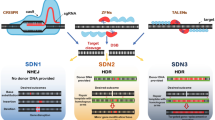
In genome-editing technology, double-stranded breaks (DSBs) are generated at a specific genome location using engineered nuclease (also called site-directed nuclease, SDNs). Nucleases are engineered by fusing a non-specific nuclease domain with a sequence-specific DNA-binding domain to precisely locate and break DNA double strand specifically. Based on the mechanism involved in using SDNs, genome editing in plant cells has been categorized into three types: SDN1 generates site-specific random mutations without the help of a donor template for DNA repair (NHEJ); SDN2 generates distinct and defined site-specific mutations using a repair template (HR); and SDN3 involves the site-specific insertion of long stretches of DNA with a repair template having flanking identical sequences (HR) or integration of a donor DNA sequence without homology (NHEJ) (Hilscher et al. 2017).
Nevertheless, CRISPR technology involves identifying the PAM sequence in the nucleotide/stretch of nucleotide to be targeted, followed by synthesizing a single gRNA (sgRNA) to clone into a suitable binary vector. The RNA polymerase III promoters (AtU6 for Arabidopsis; TaU6 for wheat; OsU6 or OsU3 for rice, etc.) are generally used in the cassette to express Cas9 and gRNA genes and eventually introduced into the host cell, followed by screening and validation of edited events.
However, there have been several bottlenecks in this technology. Target specificity, efficiency, use in a non-transgenic way, use in perennial crops, ideal delivery system, etc., are major challenges hindering it from reaching its potential. To use it in agricultural crops, transgene-free editing is essential, where proper gene containment is a challenge.
Moreover, plants generated through conventional CRISPR-based gene-editing techniques come under tight regulation of transgenic and considered GMOs as recombinant DNA constructs involved in this process (Sprink et al. 2016), thereby restricting the use of editing technology for crop improvement and agriculture as a whole.
Plant breeders across the globe are therefore focusing on improving this powerful technology with various amendments to generate foreign DNA-free agri-products. In this article, an exhaustive discussion on technological intervention has been elaborated to allow readers to understand the need and scope of CRISPR as non-GM technology in agricultural biotechnology.
Research on DNA-free editing
Thirty-six research publications on DNA-free CRISPR-based genome editing in plants were analyzed and studied thoroughly. To identify the research articles, we searched the Web of Science and Google Scholar as indexed search engines using the keywords ‘DNA-free’, ‘plant’, and ‘genome editing’ on 20th October 2022. The results obtained from both engines were merged following the removal of duplication and non-research articles. Results suggest that the number of research articles published on this theme has increased since 2015, but there is a down-trending after 2020 (Fig. 1). This may be due to the sudden COVID-19 outbreak or difficulty in the usage of this technology, since explant regeneration protocols are difficult and yet to be standardized for most of the crops. China is the leading country in DNA-free editing research (approximately, 47% of total research publications) after the USA (approximately 22%).
Moreover, several innovative concepts for foreign DNA-free approach have been developed during 2019–2022. As analyzed, most of the research works belonged to ribonucleoprotein (RNP)-mediated genome editing to achieve DNA-free status until 2020, but in later periods there were in addition many other modified and novel technologies in this arena. There have been a few studies on RNA-based editing, base editing, and nanoparticle usage. However, the number of publications is limited to a few crops only, indicating the urgent need to disseminate the novel advanced strategic tools among researchers globally to get more out of implied sciences.
The DNA-free genome-editing system
DNA-free genome editing can be defined as altering genomic sequences without keeping a trace of foreign DNA footprint in the target organism. However, ‘transgene free’ does not necessarily mean the process is foreign DNA free; rather, the ultimate product is free of foreign nucleic acid insertion. It is a well-known fact that, for sexually propagated crops, genetic segregation is quite helpful when the desirable trait is being inserted, and undesired traits can be eliminated in successive generations easily. However, in asexually propagated crops, the incorporation of a trait often leads to the insertion of an undesirable fragment of DNA. Additionally, unwanted mutations, due to illegitimate recombination events, in plant transformation enhance the chances of product rejection, though it mimics natural mutation events. So, it is a need of the hour to challenge the global demand of food supply through quality agri-products using the existing robust CRISPR technology blended with modern tools to produce ample products with the trademark ‘transgene free’.
Several coupling tools and technologies have been implied on traditional CRISPR–Cas9 systems to use in a versatile format.
Traditional DNA-free approach
The CRISPR gene-editing construct, which includes both the Cas9 and gRNA cassettes, as well as selection markers such as the kanamycin-resistant gene, is incorporated in a single plasmid. The construct is then introduced into plant cells either via Agrobacterium-mediated or biolistic transformation (gene gun). Once integrated into plant chromosomes, the transgenes can produce CRISPR enzymes and gRNAs for editing target genes. The majority of the T0 plants contain the CRISPR transgenes, even though some of them are edited at the target loci.
T0 plants are usually heterozygous at the transgene locus. In the progeny of T0 plants, both transgene and edited loci segregate according to Mendelian genetics. If only one copy of the transgenes was inserted into the genome, one-fourth of the T1 plants are transgene free. Gene gun methods and Agrobacterium-mediated transformation often result in multiple transgene insertions (Fig. 2).
In addition, if the transgene locus and edited locus are linked, it would be difficult to eliminate the transgenes by genetic segregation. To obtain transgene-free and edited plants, selfing or backcrossing T0 plants to wild-type plants take an extra generation. Therefore, plants free of transgenes can be isolated from the progeny of T0 plants or backcrossed populations using genetic segregation, but the process is laborious and time-consuming. Additionally, a few other methods can also be employed to remove transgenes such as the usage of the recombinase system (Anand et al. 2019).
Preassembled CRISPR/Cas9 or RNP system
It is possible to transfect plant cells with non-integrating plasmids to deliver programmable nucleases such as Cas9. Nevertheless, transfected plasmids are degraded in host cells by endogenous nucleases, resulting in small DNA fragments inserted in both on-target and off-target sites. Therefore, if regulatory approval is required, this approach might not be appropriate for agricultural crops.
Delivering preassembled Cas9 protein–gRNA ribonucleoproteins (RNPs) to plant cells rather than plasmids encoding these components may reduce the likelihood of recombinant DNA being inserted into the host genome (Fig. 3). Further, RNA-guided endonucleases (RGEN)- RNPs cleave chromosomal target sites immediately following transfection and are rapidly degraded by endogenous proteases. In whole regenerated plants, it may reduce the frequency of mosaicism and off-target effects. Because it is not necessary to optimize codon usage or to find promoters that will express Cas9 and gRNA when using protein-and-RNA-only systems, preassembled RGEN-RNPs could enable genome editing to be applied to a wide range of crop species.
The process involves mixing purified Cas9 protein with an excess of gRNAs targeting the desired gene(s) of a plant species in vitro to form preassembled RNPs. RGEN-RNPs are then incubated in the presence of polyethylene glycol (PEG) with protoplasts isolated from the concerned species. The T7 endonuclease I (T7E1) assay and targeted deep sequencing can both be used to measure mutation rates in transfected cells. It is always expected that a change in the three nucleotides upstream of a 5’-NGG-3’ protospacer-adjacent motif (PAM) will occur (Woo et al. 2015).
Mutations caused by RGENs were detected 24 h after transfection, suggesting they cut target sites immediately after transfection and induce mutation before a full cell division cycle is completed. The germline transmission of the targeted mutation with a higher frequency (> 45% in the case of lettuce) and lower off-target mutation frequency (off-target in 7nt mismatch) is indicative of its use in DNA-free agricultural crop production.
CRISPR-Cpf1 system
CRISPR-Cpf1 (CRISPR from Prevoltella and Francisella1), a ribonuclease, is also an excellent tool for DNA-free genome editing. It differs from the Cas9 system in several ways. This enzyme recognizes PAM sequences with T-rich (5'-TTTN-3’) at the 5'-end of DNA sequences. A single crRNA guides Cpf1, and no trans-acting crRNA is required. One of the Cpf1 family proteins is LbCpf1 from the Lachnospiraceae bacterium ND 2006, while AsCpf1, from the Acidaminococcus sp. BV3L6, acts more effectively.
Cpf1 crRNAs, however, are shorter than Cas9 sgRNAs by 60 nucleotides, allowing us to use chemically synthesized crRNAs. The deletions induced by Cpf1 are larger than those induced by SpCas9. Lastly, the Cpf1 cleavage pattern may facilitate NHEJ-mediated insertion of donor DNAs, which would yield SDN1 and SDN2 editing.
The advantage of using RNPs is that off-target effects and cytotoxicity associated with DNA transfection are significantly reduced. To transfect the protoplast, the recombinant LbCpf1 and AsCpf1 proteins mixed with crRNAs (~ 1:6 molar ratio of Cpf1:crRNA) in the presence of PEG solution has been reported recently. It was found that Cpf1-induced mutation was majorly due to deletions of several nucleotides (Kim et al. 2017), and no off-target mutation and foreign DNA trace were detected. This makes it an effective tool for editing genomes in agriculture without using DNA.
The only drawback of this system is that Cpf1–crRNA complexes could induce mutations at one- or two-base mismatch sites, but do not tolerate four or more mismatches. Therefore, care should be taken while designing gRNA to avoid the off-target effect. crRNAs need to be selected without allowing three nucleotide mismatches based on the entire homology search in the current reference genome of targeted crop species, except the target sites.
Base editing system
Genome editing can be refined through precise and predictable point mutations to decipher natural genomic variations and improve crop breeding. However, DNA double-stranded break (DSB) repair mechanism is either error prone (NHEJ) or demands a donor DNA fragment insertion (HR), which is more challenging when the inheritance of the introduced mutations and the removal of transgenes are vital issues that need to be addressed.
Recently, base editors have been developed using either an adenine or a cytidine deaminase fused to Cas9 nickase (nCas9; Cas9 having no nuclease activity), resulting in a C-to-T or an A-to-G substitution, respectively, without introducing DSB. nCas9/deaminase fusion is driven to the target locus via single-guide RNA molecules (sgRNA), which enable deamination on the non-complementary strand of DNA (Fig. 4).
A cytidine base editor (CBE), called Target-AID (target activation-induced cytidine deaminase), is a powerful tool used to edit target sequences. So far, several crops have been edited using this system with high mutation frequency (~ 70%).
For example, by transiently expressing a cytosine base editor and a guide RNA in protoplasts, a mutation was induced in the carrot genome (Daucus carota) (Meyer et al. 2022). Afterward, the protoplasts are cultured to produce somatic embryos, which later develop into carrot plants. With this approach, the plasmid DNA encoding the gene-editing machinery is not stably incorporated into the plant cells. Instead, editing occurs during the first few days following transformation before the introduced plasmid DNA is degraded and lost from the cells.
The only demerit of this system is the involvement of plant transformation using the tedious Agrobacterium-mediated vector system, where crRNA and recombinant nCas9 are cloned prior to being inserted. Additionally, this system relies on Mendelian segregation and therefore seeks time and selection pressure to confirm the non-transgenics.
However, for heterozygous and polyploid crops, this tool is very useful (e.g., potato), though the adenine base editor displays a much cleaner base edition than cytidine base editors (Veillet et al. 2019). As per the mutation type reported, base conversion is mainly C-to-G and C-to-T, while C-to-A is much less frequent using this system. This system critically relies on a selectable marker (e.g., herbicide-tolerant plants) with simultaneous multiplex nucleotide substitutions for the desired trait. Additionally, the feasibility of multiple-base editing or co-base editing broadens the scope of using this system (Shimatani et al. 2018).
Transient editing
CRISPR/Cas9 ribonucleoproteins are delivered to the cell of a plant through particle bombardment, or RNPs are delivered to protoplasts without the need for stable integration of the CRISPR/Cas9 genes into the host-plant genome. Both are successful systems for transgene-free crop production. In some species, however, the possibility of full-plant regeneration is limited by using protoplasts and biolistics.
The use of Agrobacterium to transiently express the Cas9 and sgRNA genes in tobacco without integrating foreign DNA was reported to be successful. This could be applied in perennial crop species, where the above-mentioned two approaches are not feasible. There is, however, a need for a visible marker in this system, since a large number of mutants are reported to be screened (Chen et al. 2018a).
By avoiding the use of chemical selection such as kanamycin, this approach is regeneration friendly. Without a chemical selection agent, plants regenerate even faster after infection with Agrobacterium, which increases the production of mutant calli and shoots.
This system has the disadvantage of generating very few transgene-free and edited T0 plants. However, this approach still yields more edited lines than the RNP-mediated system, where all T0 plants are transgene free, but very few are edited.
Use of the viral system
To achieve single, multiplex mutagenesis and chromosome deletions at high frequency (~ 90%), a plant negative-strand RNA virus-based vector is suitable for DNA-free in planta delivery of the entire CRISPR–Cas9 cassette to plant cell. In contrast to positive stranded viral replicons where size insertion and movement within the host are major constraints, this is a convenient, highly efficient, and cost-effective approach in which a large CRISPR–Cas9 cassette can be transferred.
To exploit the use of plant viral vector for non-transgenic delivery of CRISPR–Cas9 reagents, the Streptococcus pyogenes Cas9 and gRNA sequences were inserted between the N and P genes of the SYNV genome in which the gRNA sequence was flanked by two pre-tRNAGly sequences resulting in tRNA SYNV–tgtRNA–Cas9 construct (Ma et al. 2020). Multiple gRNAs can be used in the construct to target multiple sequences at a time.
Mutant plants regenerated from infected tissues are considered non-transgenic, because RNA viruses like SYNV (Sonchus yellow net rhabdovirus) do not integrate into host chromosomes during replication. SYNV facilitates the direct delivery of CRISPR–Cas9 reagents into intact plants, allowing for the regeneration of diverse plant tissues suitable for regeneration unlike previously developed DNA-free delivery methods requiring the isolation of specialized plant cells or tissues. In addition to the extraordinary stability of the vector, the progeny viruses can also be mechanically passed from plant to plant, thereby avoiding the use of Agrobacterium pathogens altogether (Fig. 5).
Major limitations include the possibility of off-target mutations when there is a mismatch of two to five nucleotides, and the long-term stability of the construct inside the host cell increases the mutation probability. In general, viral delivery systems are limited by the host range associated with a given virus species. However, reverse-genetics tools are becoming more readily available for similar viruses and the effort to generate or identify a more convenient viral vector system is also being undertaken (Zhou et al. 2019).
Ribozyme-mediated guide RNA production and fluorescence-based technology
Ribozyme-based sgRNA production opens a door for conducting more sophisticated gene-editing experiments that would not be possible by using RNA Pol III promoter-based technology (Fig. 6). Because primary transcripts from the Ribozyme1-gRNA-Ribozyme2 construct are automatically processed to release the sgRNA molecule, we can use any promoter to produce sgRNA. Cell/tissue-specific promoters would enable gene editing in specific cells.
This procedure involves flanking the desired sgRNA molecule with ribozymes and RNAs with enzymatic activity, at both 5' and 3' ends to facilitate self-catalyzed cleavage to release the mature sgRNA for gene editing. Due to the presence of ribozymes at both ends, any post-translational modifications such as capping or polyadenylation will not affect the designed sgRNA. Therefore, any promoter can be used when designing a CRISPR–Cas construct, as opposed to a typical CRISPR–Cas construct that requires a promoter for polymerase III (He et al. 2017).
Using this system has the advantage that no additional bases are added during the transcription of sgRNA, making the process more accurate and reducing the number of off-target mutations.
Additionally, seeds can be selected with integrated gRNA–Cas construct using any fluorescent-based marker, such as mCherry fluorescence as a proxy for the presence of the CRISPR/Cas9 construct, instead of using any selectable marker like kanamycin or hygromycin. A breeder can eliminate edited T0 seeds showing fluorescence and select only non-fluorescent seeds for generating transgene-free plants and follow the same in successive generations to further consider the heritability of an edited trait. Moreover, foreign DNA-free plants are easily identified at the seed stage, eliminating the laborious work of planting (Aliaga-Franco et al. 2019).
Recently, it has been reported that a modified and efficient promoter, OsU3-tRNA promoter, in combination with CRISPR/Cas9 system contributed to the highest mutagenesis efficiency that increased sgRNA expression levels over the AtU6-tRNA and AtU6 promoters, (Zhang et al. 2022). They optimized the existing tobacco CRISPR/Cas9 system by using the OsU3-tRNA promoter combination instead of AtU6 and fusing an AtUb10-Ros1 expression cassette in T-DNA for monitoring the transgene events. The new vector was named pOREU3TR. To obtain stably transmissible mutations in tobacco generated by CRISPR/Cas9-mediated genome-editing technology, it is necessary to segregate the CRISPR/Cas9 construct. However, it is laborious and inefficient to isolate the Cas9-free mutants in the transforming generations by the traditional PCR method. Therefore, they inserted a dark red tag (Ros1) driven by the AtUb10 promoter into OsU3-tRNA to obtain a visual screen of mutants. When plasmids containing the pOREU3TR-target gene unit were transformed into tobacco, anthocyanin accumulated in the callus, leaves, stem, root, and flowers, producing a visible dark red color, which can facilitate the detection of transgenic events in callus and plants. All of the T0 plants that displayed dark red color contained the Cas9 expression cassette, which was confirmed by PCR analyses later.
Therefore, fluorescent-based screening is very useful in shortening the time requirement of non-transgenic selection.
Nanoparticle-based system
Biotechnological applications are hampered by the recalcitrance of economically important plant species. By contrast, nanoparticles (NPs) have the ability to deliver biomolecules to plants for genome editing and have emerged as a crucial driver for improving plant transformation efficiency. They include inorganic NPs, carbon-based NPs, silicon-based NPs, and polymeric NPs. DNA is the only genetic cargo that inorganic NPs (gold, silver, iron oxide) can transport inside plants.
A different form of CRISPR/Cas9 has been used in combination with various nanoparticles, viz., nanocapsules, gold nanoparticles, hydrogels, peptide-based NPs, DNA nanoclew, polymeric NPs, unilamellar liposomes NPs, multilamellar liposomes, and magnetic NPs where they act as carriers for delivery into the host cell to facilitate incorporation of target genes with encoded nanoparticles into the cells. Plants can uptake these combined nanoparticles to make the desired alteration in the target gene sequence.
Zhao et al. (2017) studied genetic modification in pollen using magnetic NPs exogenous DNA delivery. Positively charged magnetic nanoparticles (MNPs) are used in pollen magnetofection technology to bind with electrically negative DNA to form the MNP–DNA complex. A magnetic field is applied after MNP–DNA complexes are blended with pollen, directing the complexes into the pollen via apertures before pollination occurs. With this method, the DNA functions and viability of the pollen are maintained. Furthermore, the process is easy to perform, requires no equipment, and is capable of delivering multiple genes. They revealed genetic transformation by pollen magnetofection by maintaining pollen viability, resulting in the development of insect-resistant transgenic seeds of cotton. They reported that pollen mangnetofection is an advanced technology with high-efficiency in-field operations (Zhao et al. 2017). This system is culture free and genotype independent (Fig. 7). In addition, it is simple and fast as no regeneration time is required and is capable of multi-gene transformation.
The direct uptake of nanoparticles such as quantum dots, metal/metal oxide NPs, silica NPs, and carbon nanotubes has also been reported earlier in various crops, suggesting its efficiency to generate transgene-free crops. Gold nanoparticles (AuNP), when compared to other nanoparticles, possess excellent physicochemical stability and excellent biocompatibility. CRISPER–Cas9 modes can be delivered by primarily gold-based nanomaterials and lipid NPs, AuNP and AuNC (Chen et al. 2019; Vats et al. 2022).
Nanoparticles can potentially deliver gene-editing cargo to any plant cells including meristematic cells (Sanzari et al. 2019; Khan et al. 2019). Delivery of gene-editing reagents through nanoparticles into meristematic cells can potentially generate chimerically edited plants. Transgene-free and edited plants may be regenerated from the edited tissue through tissue culture or from the propagation of cuttings. A recent exciting report indicates that plasmid-coated carbon dots can be delivered into plant cells by foliar application (spraying on) and that Cas9/gRNAs produced by this method successfully edited target genes (Doyle et al. 2019). This new method can be potentially extended to other plants, offering a simple, fast, and inexpensive method for editing plant genomes.
Nevertheless, this approach also has several unanswered questions. First, optimization of the correct dose/concentration of nanoparticle or functionalized material to be used for coating is very important because of high reactivity and less stability. Second, biosafety knowledge of the NPs being used for gene delivery in a particular plant/tissue should be clear. Third, the binding affinity of cargos and nanoparticles decides the performance of transformation efficiency. Fourth, the mechanism of delivery and the fate of nanoparticle and/or functionalized material should be studied to understand the biocompatibility/biosafety/toxicity of these materials.
TKC and CASE toolkit approach
Transgene killer CRISPR (TKC) technology relies on spatial–temporal expressions of suicide cassettes containing p35S:: Cytoplasmic Male Sterility 2 (CMS2) and pREG2::BARNASE to kill all transgene-containing sperms and embryos, respectively (He et al. 2018), to select transgene-free plants (Fig. 8).
The researchers placed the bacterial BARNASE gene, which encodes a toxic protein with nuclease activity, under the control of the rice REG2 promoter, known to be expressed during early embryonic development, into the regular CRISPR/Cas9 plasmid pCXUN-Cas9. The REG2-BARNASE cassette kills any embryos that contain the transgenes. Rice male gametophyte-specific lethal protein CMS2, which is also called ORFH79, disrupts mitochondria functions during male gametophyte development and causes male sterility. It was used in the expression cassette to kill transgene-containing male gametophytes.
Recently, Customized Assembly and Simplified Editing (CASE) toolkit developed in rice (Oryza sativa) combines the above TKC technology with advancement in multiplex gene editing (Fig. 8). This approach provides an easy and efficient way to obtain transgene-free gene-edited plants for multiple genes in T1 generation (Chen et al. 2018b; Liu et al. 2022).
The CASE toolkit consists of a set of four gRNA cloning vectors that contain either U3, U6a, U6b, or U6c small non-coding RNA (snoRNA) promoters, and a TKC-MCS-U3 gene-editing backbone vector. The former allows a one-step assembly of independent gRNAs to create customized combinations of gRNA cassettes with compatible restriction sites in flanking regions. Then, the combinations of gRNA cassettes for multiple editing targets are transferred to the latter using restriction sites at MCS. Alternatively, chemically synthesized gRNA cassettes, spaced with self-splicing tRNA, can also be assembled to the TKC-MCS-U3 backbone for multiplex editing.
An improved TKC technology in rice by replacing the CaMV35S promoter has been reported (Yubing et al. 2019). They replaced the CaMV35S promoter with the rice ACTIN1 promoter, as CamMV35S is relatively weak in monocots and especially weak in floral organs, particularly in the microspore cells, which are strong and constitutive. They also used a pollen-specific promoter OsGEX2 promoter to drive CMS2 expression in pollen cells. Both strategies led to successful isolation of transgene-free and edited rice plants within a single generation.
This technological intervention has huge potential in transgene-free genome editing using the advantage of TKC technology of non-transgenics rejection, more specifically in multiplex editing with a high frequency of editing and selective removal of non-transgenics in rice. However, the scope is yet open to other crops.
Promoter editing
In 2020, scientists from China successfully implied the CRISPR–Cas9 system to knock out the distinct region of promoter of the xa13 gene through deletion using two specific gRNA. The xa13 gene has a role in bacterial blight disease susceptibility as well as pollen fertility. So, targeting the CDS of this gene was not possible to make it resistant to bacterial blight. However, the deletion of the promoter sequence would cause the Xa13 gene to lose its ability to be induced by bacterial blight, thus making the rice lack this fragment resistant to bacterial blight without losing pollen fertility (Li et al. 2020).
This minor modification was successfully executed in the promoter and the line developed is considered non-transgenic, because in addition to eliminating the exogenous DNA transgene fragments, editing the promoter region simply changes the expression pattern of the gene without producing excess mRNA or protein, in contrast to the conventional editing of gene coding regions to generate frame‐shifted or erroneous mRNA or protein. Editing the promoter region produces transgene‐free rice. A double‐sgRNA site‐directed mutation was directed to the DNA sequence deletion; it was easy to use PCR to identify whether the mutation site was a homozygous, heterozygous, or no‐deletion mutation. It greatly reduced the sequencing workload and improved the accuracy of selecting mutant plants. The two sgRNA‐mediated deletion mutations are relatively stable compared to the single‐target‐induced mutation, and the mutation result is predictable.
In 2019, another excellent strategy was reported in maize crop, where editing through haploid induction to get transgene-free lines was successful. This technique is also called Hi-Edit technology (Kelliher et al. 2019). The Hi-Edit method involves first transformation of the CRISPR/Cas9 construct into NP2222, a common transformation line. To select F2 individuals homozygous for both the haploid-inducing gene and the Cas9 insertion, Cas9+ progenies from regenerated plants were crossed with a native haploid-inducer line, RWKS. Elite inbred lines were fertilized with pollen from these F2 individuals. In the descendant haploid progenies, the transgene-free mutant of interest was identified.
Choice of explant and other limitations
Though the exclusion of foreign DNA integration is fascinating, there are several limitations in transgene-free CRISPR-based editing technologies. A few are discussed below:
-
(1)
Protoplast (a cell without a cell wall) is the most preferable explant in targeting using the PEG-based transformation method, but isolation and regeneration of protoplast are challenging tasks and sometimes not feasible in certain crops such as perennial crops. Moreover, protoplast is very susceptible to mechanical injury and therefore sometimes may lead to poor regeneration.
-
(2)
Using particle bombardment, editing is successful in many crops, but also has several limitations such as low transformation efficiency than agro-based transformation, tissue damage, requirement of a higher quantity of DNA, etc.
The reproducible robust regeneration process is the critical parameter while the transfection method and explant selection are concerned. Though different sets of strategic establishments have been trialed so far, yet a challenging task for plant biotechnologists. A simplified view of DNA-free editing is shown in Fig. 9.
Role of transgene-free approaches of CRISPR-based genome editing in stress tolerance of crop plants
Biotic and abiotic stress tolerance can be achieved by targeting the stress-sensitive genes or their cis-regulatory elements. For example, in the case of rice, SWEET14 gene was targeted to induce resistance against blight disease (Blanvillain-Baufumé et al. 2017). Though transgene-free targeted genome editing-based stress tolerance crop improvement is a new concept, it has already been established as a successful technology in yield-based traits. Previously, many attempts were made to either transiently overexpress resistance genes, but recent advances have made targeted editing of genes for desired traits more feasible. The above-mentioned approaches can be used to integrate biotic and abiotic stress resistance traits into crop breeding programs. New research avenues have undoubtedly opened up for speedy improvement of important and complex traits such as drought, salinity, and heat tolerance.
Regulatory concerns and prospects
Genome editing and natural mutations are usually indistinguishable from each other as far as base substitution/indel is concerned (Hartung and Schiemann 2014; Ran et al. 2017). As endonucleases are degraded within a few hours, their mode of action will be inactive in the targeted cell. The plant regenerated thus will have no transgene inserted within the genome (Liang et al. 2017). If we focus on the world’s GMO acceptance scenario, different prospects may be seen. The three agencies responsible for regulating GMOs in the USA, i.e., the Department of Agriculture, The Food and Drug Administration, as well as the Environmental Protection Agency (EPA), evaluate genetically altered organisms by taking the end product (not the procedure followed) into account. Therefore, plants with permanently incorporated transgenes are considered GMOs. However, Europe focuses on the transformation process. Thus, in 2019, the European Court of Justice (ECJ) decided that products derived from genome-editing processes will also fall under the strict regulatory framework of GMOs, leading to latent trade issues and hindering novelty as per WTO (World trade organization 2018). Recently, Argentina, Brazil, Chile, the USA, Canada, Paraguay, and Uruguay declared genome-editing-friendly regulations and to consider genome-edited plants equivalent to conventionally bred lines. In India (2022) also, CRISPR technology falling under the categories of SDN-1 and SDN-2, will be exempted from biosafety regulations.
The progress in DNA-free plant genome editing is directed toward a modification based on reduced off-target and increased transformation efficiency along with good regeneration efficiency of explants. More than a dozen crops have been (enlisted in Table 1) trialed with this approach to improve different traits, but a robust protocol for transfection yielding effective regeneration is yet to be achieved. Researchers need to devote greater focus to achieve better efficacy of this technology.
To feed the ever-expanding global population (by 2050, the human population will reach approx. ten billion) and to beat the continuous challenge of climate change and biotic stresses, global food production needs to rise by 60–100% (FAOSTAT, 2016) with limited natural resources, including genetic resources. So, international harmonization of the regulations implies that DNA-free genome editing has become critical to understand and safeguard the invention perspective of genome-edited plants.
Conclusion
Traditionally, genome editing involves integrating editing components (gRNA and Cas construct) into the host genome. Even if the DNA construct gets degraded, the resulting fragments may randomly be integrated illegitimately wherever a DSB happens in the genome. Additionally, any homology sequence available in the genome could produce unwanted effects. On the other hand, continued expression of genome-editing components intensifies the off-target effects. Therefore, genome editing without permanent insertion of foreign DNA trace is the leading-edge technology, generating genetically edited crops with minimal risk of unwanted off-target mutations and meeting existing and future agriculture demands from a scientific and regulatory perspective. Two direct transfection methods, viz., protoplast-mediated transformation and particle bombardment, have both been successfully used in a few crop species for DNA-free genome-editing technology. Nevertheless, unfortunately, efficient regeneration protocols are not available for most crops, indicating an emerging need to bring down the gap to use this prevailing technology in a world-sustainable agricultural scenario.
Abbreviations
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- DBD:
-
DNA-binding domain
- DBP:
-
DNA-binding proteins
- DR:
-
Direct repeat
- DSB:
-
Double-stranded break
- ECJ:
-
European Court of Justice
- GBSS:
-
Granule-bound starch synthase
- GMO:
-
Genetically modified organisms
- HR:
-
Homologous recombination
- NC:
-
Nanocapsule
- NHEJ:
-
Non-homologous end joining
- NSV:
-
Negative-stranded virus
- PAM:
-
Protospacer-adjacent motif
- PEG:
-
Polyethylene glycol
- WTO:
-
World Trade Organization
References
Aliaga-Franco N, Zhang C, Presa S et al (2019) Identification of transgene-free CRISPR-edited plants of rice, tomato, and Arabidopsis by monitoring DsRED fluorescence in dry seeds. Front Plant Sci 10:1150. https://doi.org/10.3389/FPLS.2019.01150/BIBTEX
Anand A, Wu E, Li Z et al (2019) High efficiency agrobacterium-mediated site-specific gene integration in maize utilizing the FLP-FRT recombination system. Plant Biotechnol J 17:1636–1645. https://doi.org/10.1111/PBI.13089
Blanvillain-Baufumé S, Reschke M, Solé M et al (2017) Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J 15:306–317. https://doi.org/10.1111/PBI.12613
Chen L, Li W, Katin-Grazzini L et al (2018a) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res 5:13. https://doi.org/10.1038/S41438-018-0023-4/42567482/41438_2018_ARTICLE_23.PDF
Chen L, Li W, Katin-Grazzini L et al (2018b) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res. https://doi.org/10.1038/S41438-018-0023-4
Chen G, Abdeen AA, Wang Y et al (2019) A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol 14:974–980. https://doi.org/10.1038/S41565-019-0539-2
Doyle C, Higginbottom K, Swift TA et al (2019) A simple method for spray-on gene editing in planta. bioRxiv. https://doi.org/10.1101/805036
Hartung F, Schiemann J (2014) Precise plant breeding using new genome editing techniques: opportunities, safety and regulation in the EU. Plant J 78:742–752. https://doi.org/10.1111/TPJ.12413
He Y, Wang R, Dai X, Zhao Y (2017) On improving CRISPR for editing plant genes: ribozyme-mediated guide RNA production and fluorescence-based technology for isolating transgene-free mutants generated by CRISPR. Prog Mol Biol Transl Sci 149:151–166. https://doi.org/10.1016/BS.PMBTS.2017.03.012
He Y, Zhu M, Wang L et al (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Cell Com. https://doi.org/10.1016/j.molp.2018.05.005
Hilscher J, Bürstmayr H, Stoger E (2017) Targeted modification of plant genomes for precision crop breeding. Biotechnol J. https://doi.org/10.1002/BIOT.201600173
Ishino Y, Shinagawa H, Makino K et al (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433. https://doi.org/10.1128/JB.169.12.5429-5433.1987
Jinek M, Chylinski K, Fonfara I et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (1979) 337:816–821. https://doi.org/10.1126/SCIENCE.1225829
Kelliher T, Starr D, Su X et al (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37:287–292. https://doi.org/10.1038/S41587-019-0038-X
Khan I, Saeed K, Khan I (2019) Nanoparticles: properties, applications and toxicities. Arab J Chem 12:908–931. https://doi.org/10.1016/J.ARABJC.2017.05.011
Kim H, Kim ST, Ryu J et al (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:1–7. https://doi.org/10.1038/ncomms14406
Li C, Li W, Zhou Z et al (2020) A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol J 18:313. https://doi.org/10.1111/PBI.13217
Liang Z, Chen K, Li T et al (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:1–5. https://doi.org/10.1038/ncomms14261
Liu JL, Chen MM, Chen WQ et al (2022) A CASE toolkit for easy and efficient multiplex transgene-free gene editing. Plant Physiol 188:1843–1847. https://doi.org/10.1093/PLPHYS/KIAB573
Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants 6:773–779. https://doi.org/10.1038/s41477-020-0704-5
Malnoy M, Viola R, Jung MH et al (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904. https://doi.org/10.3389/FPLS.2016.01904/BIBTEX
Meyer CM, Goldman IL, Grzebelus E, Krysan PJ (2022) Efficient production of transgene-free, gene-edited carrot plants via protoplast transformation. Plant Cell Rep 41:947–960. https://doi.org/10.1007/S00299-022-02830-9/TABLES/2
Murovec J, Guček K, Bohanec B et al (2018) DNA-free genome editing of brassica oleracea and B. Rapa protoplasts using CRISPR-cas9 ribonucleoprotein complexes. Front Plant Sci 871:1594. https://doi.org/10.3389/FPLS.2018.01594/BIBTEX
Ran Y, Liang Z, Gao C (2017) Current and future editing reagent delivery systems for plant genome editing. Sci China Life Sci 60:490–505. https://doi.org/10.1007/S11427-017-9022-1
Sanzari I, Leone A, Ambrosone A (2019) Nanotechnology in plant science: to make a long story short. Front Bioeng Biotechnol. https://doi.org/10.3389/FBIOE.2019.00120
Shimatani Z, Fujikura U, Ishii H et al (2018) Inheritance of co-edited genes by CRISPR-based targeted nucleotide substitutions in rice. Plant Physiol Biochem 131:78–83. https://doi.org/10.1016/J.PLAPHY.2018.04.028
Sprink T, Eriksson D, Schiemann J, Hartung F (2016) Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts. Plant Cell Rep 35:1493–1506. https://doi.org/10.1007/S00299-016-1990-2/FIGURES/3
Svitashev S, Schwartz C, Lenderts B et al (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature Communications 2016 7:1 7:1–7. https://doi.org/10.1038/ncomms13274
Subburaj S, Chung SJ, Lee C et al (2016) Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep 35:1535–1544. https://doi.org/10.1007/S00299-016-1937-7
Vats S, Kumawat S, Brar J et al (2022) Opportunity and challenges for nanotechnology application for genome editing in plants. Plant Nano Biol 1:100001. https://doi.org/10.1016/J.PLANA.2022.100001
Veillet F, Perrot L, Chauvin L et al (2019) Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci 20:402. https://doi.org/10.3390/IJMS20020402
Woo JW, Kim J, Kwon S et al (2015) DNA-free genome editing in plants with preassembled CRISPR–Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164. https://doi.org/10.1038/nbt.3389
Yubing H, Min Z, Lihao W et al (2019) Improvements of TKC technology accelerate isolation of transgene-free CRISPR/Cas9-edited rice plants. Rice Sci 26:109–117. https://doi.org/10.1016/J.RSCI.2018.11.001
Zhang Y, Liang Z, Zong Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nature Communications 2016 7:1 7:1–8. https://doi.org/10.1038/ncomms12617
Zhang J, Mi Q, Yang W et al (2022) Highly efficient transgene-free plant genome editing in tobacco using an optimized CRISPR/Cas9 system pOREU3TR. Plant Sci. https://doi.org/10.21203/rs.3.rs-1637574/v1
Zhao X, Meng Z, Wang Y et al (2017) Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat Plants 3:956–964. https://doi.org/10.1038/S41477-017-0063-Z
Zhou X, Sun K, Zhou X et al (2019) The matrix protein of a plant Rhabdovirus mediates superinfection exclusion by inhibiting viral transcription. J Virol 93:680–699. https://doi.org/10.1128/JVI.00680-19/ASSET/4B05DC68-A057-499D-8E3E-B6ADA94FB524/ASSETS/GRAPHIC/JVI.00680-19-F0007.JPEG
Zong Y, Wang Y, Li C et al (2017) Precise base editing in rice, wheat and maize with a Cas9- cytidine deaminase fusion. Nature Publishing Group 35:. https://doi.org/10.1038/nbt.3811
Funding
The authors have no funding source.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest associated with the publication. There are no known competing financial interests or personal relationships that could have influenced the work reported in this article.
Human and animal participants
This research involves no human/or animal participants.
Informed consent
Informed consent was taken from all the authors. The authors also declare that all data were generated in-house.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhattacharjee, S., Bhowmick, R., Kant, L. et al. Strategic transgene-free approaches of CRISPR-based genome editing in plants. Mol Genet Genomics 298, 507–520 (2023). https://doi.org/10.1007/s00438-023-01998-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-01998-3