Abstract
The goal of the study was to design a single tube PCR test for detection and differentiation of Babesia species in DNA samples obtained from diverse biological materials. A multiplex, single tube PCR test was designed for amplification of approximately 400 bp region of the Babesia 18S rRNA gene. Universal primers were designed to match DNA of multiple Babesia spp. and to have low levels of similarity to DNA sequences of other intracellular protozoa and Babesia hosts. The PCR products amplified from Babesia DNA isolated from human, dog, rodent, deer, and tick samples were subjected to high-resolution melting analysis for Babesia species identification. The designed test allowed detection and differentiation of four Babesia species, three zoonotic (B. microti, B. divergens, B. venatorum) and one that is generally not considered zoonotic—Babesia canis. Both detection and identification of all four species were possible based on the HRM curves of the PCR products in samples obtained from the following: humans, dogs, rodents, and ticks. No cross-reactivity with DNA of Babesia hosts or Plasmodium falciparum and Toxoplasma gondii was observed. The lack of cross-reactivity with P. falciparum DNA might allow using the assay in endemic malaria areas. The designed assay is the first PCR-based test for detection and differentiation of several Babesia spp. of medical and veterinary importance, in a single tube reaction. The results of the study show that the designed assay for Babesia detection and identification could be a practical and inexpensive tool for diagnostics and screening studies of diverse biological materials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Babesia spp. are tick-transmitted protozoan hemoparasites, considered to be the second most commonly found parasites in the blood of mammals (Schnittger et al. 2012). Infections with Babesia can be fatal in humans, cattle, and companion animals (Schnittger et al. 2012). Babesiosis causes economic loss in animal production in the USA, Australia, and South America (Canever et al. 2014). Furthermore, Babesia infections pose a public health threat since the parasite can be transmitted not only through tick bites but also by transfusion of blood and blood products. However, there still are no standardized procedures of screening blood for Babesia even in such developed countries as the USA, where cases of babesiosis related to blood transfusion have been recorded (Bish et al. 2015; Leiby 2006; Levin and Krause 2016).
Four Babesia species and a few strains with uncertain taxonomic position have been recognized as pathogenic for humans so far. In North America, the main causative agent of human babesiosis is B. microti whereas in Europe, most of the described human autochthonous infections were caused by B. divergens and B. venatorum (Haapasalo et al. 2010; Zintl et al. 2003). In Asia, the public health risk associated with B. microti and B. venatorum infections has been recognized recently (Zhou et al. 2013, 2014). There are no official recommendations on laboratory diagnosis of Babesia infection both in the USA and in European countries (Hildebrandt et al. 2013; Levin and Krause 2016). Definitive diagnosis in most cases still depends upon finding of parasites on blood smear examination. The commercial serological and molecular tests used in the routine diagnostics of human babesiosis are not standardized. Most often, serological tests, designed mainly for infections with B. microti, are used in medical laboratories. However, clinicians in their practice have problems with interpretation of these test results since the positive results may correspond to: present, past, or no infection (Prince et al. 2010). False negative as well as false positive results due to cross-reactivity with other intracellular parasites such as Toxoplasma gondii and Plasmodium have also been observed in ELISA tests (Hildebrandt et al. 2013). However, the risk of confusing of Plasmodium and Babesia also occurs when microscopy is used for blood smear examination (Zhou et al. 2013).
It was shown that PCR for amplification 18S rRNA fragments can be more sensitive than microscopy for detection of B. microti infection (Teal et al. 2012; Wang et al. 2015a, b). Recently, numerous PCR-based assays for detection of Babesia, important from medical and veterinary points of view, have been developed (Li et al. 2015; Teal et al. 2012; Wang et al. 2015b). However, multiplex PCR for simultaneous detection and differentiation of several Babesia that infect humans has not been described to date, even though the need for such a tool has been expressed (Hildebrandt et al. 2013). The objective of this study was to design a single tube PCR test for detection and differentiation of Babesia species in DNA samples obtained from diverse biological materials.
Materials and methods
Biological material
Clinical and environmental DNA samples were tested: human, dog, Microtus oeconomus, Caproleus caproleus, and Ixodes ricinus (Table 1). Human clinical samples tested for malaria, babesiosis, and toxoplasmosis originated from routine diagnostics of the Diagnostic Laboratory of the Department of Parasitology NIPH-NIH. Obtaining the patients’ consent for use of their samples for research purposes is a standard procedure in the NIPH-NIH Diagnostics Laboratories. A single human blood sample positive for Babesia venatorum was collected during project NN404520038 financed by the Ministry of Science and Higher Education. The granted study was approved by the Bioethics Commission in NIPH-NIH (Sadkowska-Todys et al. 2015). The dog blood samples collected for routine diagnostics were obtained from the patients of the Antecedo (Rafal Stachurski and Katarzyna Stachurska) veterinary clinic. Blood smear examination was used by veterinarians to confirm Babesia infection. Roe deer (Caproleus caproleus) spleen sample was obtained through the courtesy of Prof. Bogumila Skotarczak from the University of Szczecin (Sawczuk et al. 2005). Microtus oeconomus and Caproleus caproleus blood samples were obtained through the courtesy of Prof. Grzegorz Karbowiak from the Polish Academy of Sciences, Warsaw. The latter samples were examined by microscopy (thick blood smears). Tick DNA samples originated from the Department’s collection.
DNA extraction
The DNA from animal blood and ticks was isolated according to the silica guanidinium protocol (Boom et al. 1999) with modifications for whole blood and arthropod samples (Masny et al. 2011, 2016). The DNA from C. caproleus spleen was isolated using DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions.
Control sample selection
The status of all samples, positive or negative for Babesia, was confirmed by PCR with universal primer pair for Babesia and Theileria amplification: Bab-GF2 (5′-GYYTTGTAATTGGAATGATGG-3′) and Bab-GR2 (5′-CCAAAGACTTTGATTTCTCTC-3′) (Bonnet et al. 2007) with minor modifications required to adjust the reaction conditions to the reagents used; Hot Star Taq DNA PCR 0.625 U polymerase, 1× PCR buffer (Qiagen), 1 μl in total volume of 25 μl, were used, the PCR started with a polymerase activation phase at 94 °C for 15 min, followed by 40 cycles at 94 °C for 60 s, 60 °C for 90 s, and 72 °C for 60 s. The PCR products were sequenced [GenBank: KP072001.1, KT844903.1, KT844904.1, KT844905.1, KT844906.1, KT844907.1, KT844908.1, KT844909.1, KT844910.1, KT844911.1, KT844912.1, KT869379.1], and the species of Babesia was determined based on the similarity to the genomic sequences deposited in GenBank.
Sequence analysis and primer design for multiplex PCR HRM
Multiple sequence alignments were performed with ClustalX2 (Larkin et al. 2007) and MAFFT algorithm using Jalview software (Waterhouse et al. 2009) and subsequently were manually edited using SeaView (Gouy et al. 2010) and Jalview software packages. The primers were designed manually, based on the alignment of the following 18S rRNA Babesia nucleotide sequences obtained from GenBank: AJ439713.1, EF458228.1, FJ944822.1, FJ944823.1 (B. divergens); AY693840.1, EF413181.1, JX417370.1, KC470047.1, KC821597.1, KF410824.1, KF410825.1, KF410826.1, KF410827.1 (B. microti); KF724377.1, KJ663730.1, KM244044.1 (B. venatorum also described as Babesia sp. EU1) and AY048113.1 (Babesia sp. MO1). The three designed primers were universal for at least seven Babesia taxons (Table 2), five of them described to infect humans: B. divergens, B. divergens-like, B. microti, B. venatorum, Babesia microti-like.
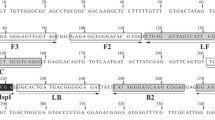
The forward primers were as follows: B-BM (5′-GAATCTAAACCCTTCCCAGAGTATC-3′), B-BDV (5′-TGACCTAAACCCTCACCAGAGTAAC-3′) and the universal reverse primer was B-rev (5′-GCTTTCGCAGTAGTTCGTCTTTA-3′). The primer pairs B-rev, B-BDV and B-rev, B-BM flanked 381 and 408 bp 18S rRNA gene fragments of B. divergens, B. divergens-like, B. venatorum and B. microti, Babesia microti-like, respectively (Fig. 1).
Multiple alignment of the 18S rRNA gene region amplified in multiplex HRM PCR. The reference sequence from B. microti is presented on the top of the alignment. The bases identical to those in the reference sequence are represented by dots in the sequences from the following: B. divergens, B. venatorum (EU1), Babesia sp. MO1. Only the bases that differentiate B. divergens, B. venatorum (EU1), Babesia sp. MO1 from the reference sequence are shown in the alignment as A, G, C, and T. The sequence of B. microti is the longest—408 bases versus 381 bases in case of sequences from the remaining three species, the gaps in those sequences are indicated by dashes on a pink background. Primer regions are indicated by arrows with the primer name above them. B-BM primer perfectly matches only the B. microti sequence. B-BDV primer perfectly matches sequences of B. divergens, B. venatorum (EU1), and Babesia sp. MO1, presented on the figure. B-BDV perfectly matches sequences of all four taxons. All four 18S rRNA gene regions, spanned by primer pairs B-BM/B-rev and B-BDV/B-rev, are polymorphic and have unique sequences
The range of primer specificity and potential of the primer cross-reactivity was estimated by NCBI BLAST search with default settings and relaxed settings (Masny et al. 2016).
Sanger sequencing of all samples sequenced in this study was performed by external service oligo.pl (Warsaw, Poland).
Multiplex PCR HRM
The reaction mix for mPCR-HRM composition was as follows: 1× concentrated SsoFast™ EvaGreen® Supermix, 0.4 μM of each primer, and 2 μl of template DNA in total volume of 25 μl. The PCR was performed in touchdown format, the initial denaturation at 98 °C for 3 min was followed by 40 cycles of three step reaction: 1st step, 98 °C for 15 s; 2nd step, touchdown from 63 to 55 °C for 30 s (gradual reduction of the temperature by 0.2 °C in every consecutive cycle); 3rd step, 72 °C for 30 s. Signal was acquired in every cycle, in green channel, at the end of incubation at 72 °C. After completion of PCR, premelt conditioning was performed for 90 s with default settings and was followed by HRM in a temperature range from 70 to 90 °C, in 0.2 °C temperature increments, with incubation for 3 s at each step.
Species differentiation
Genotype identification procedure was performed using Rotor Gene 6000 software–version 1.7 build 85 to discriminate Babesia spp. For establishing the confidence of identification, three replicates of HRM for each Babesia spp. were used. The threshold for species identification was set to 95% level of confidence. Difference plots of HRM curves were obtained with Rotor Gene 6000 software–version 1.7 build 85 to visualize inter-species differences between melting curves, each of the species was used as a reference, in separate analyses, for obtaining difference plots.
Analytical sensitivity
The detection limit of mPCR-HRM performance and detection limit studies were performed on serial dilutions of B. microti 18S rRNA fragment, amplified with the primers CRYPTOF (5′-AACCTGGTTGATCCTGCCAGTAGTCAT-3′) and CRYPTOR (5′-AATGATCCTTCCGCAGGTTCACCTAC-3′). The primer pair CRYPTOF and CRYPTOR (Herwaldt et al. 2003) spans 1770 bp DNA fragment of B. microti 18S rRNA. The region amplified by mPCR-HRM is located within the 1770 bp fragment. The amplified 1770 bp 18S rRNA fragment was subjected to agarose gel electrophoresis and isolated from the agarose gel using the Gel-Out® kit (A&A Biotechnology). The concentration of the purified DNA fragment was measured by UV spectroscopy and 7 ten-fold; serial dilutions of the PCR product in water were prepared. Seven dilutions from 5 × 106 copies/μl to 5 copies/μl were used as a template for mPCR-HRM. The mPCR-HRM reactions were performed in triplicate for each of the dilutions, and such independent reactions were performed in three independent PCR runs.
Results
In multiplex PCR with forward primers B-BDV, B-BM, and B-rev reverse primer, DNA of B. canis, B. divergens, B. venatorum, and B. microti was amplified. Application of HRM curve analysis allowed discrimination of the four Babesia species both visually and using the automatic ‘genotype’ assignment feature of the software (Fig. 2 and Fig. 3).
Differentiation of Babesia species by high-resolution melting curves. Melting curves—the plots of normalized fluorescence versus melting temperature obtained for four species. Average values of genotype assignment confidence (for the three repeats of experiments performed for each species) and the legend to the figure are presented on the graph
Babesia species discrimination by difference curves. Red B. microti, purple B. divergens, blue B. venatorum, green B. canis. The multiplex PCR products obtained from DNA of the following species were used as the references for difference curve plot. a B. microti. b B. divergens. c B. venatorum. d B. canis. Melting curves were automatically normalized and a reference curves were selected by the user. Values of each curve were automatically subtracted, by the software, from the values of the user selected reference curve and the results were presented on the plot. The more similar are two curves the closer the plotted line should be to a straight horizontal line (such as the repeats of the reference species curve on the plot of difference curves). The differences between the melting curves of the multiplex PCR products are emphasized on difference plots and observed as the differences in the shapes of the difference curves. Such curve shape differences correspond to the differences at the level of nucleotide sequence (only those sequence alterations that cause DNA melting temperature change). Differentiation of all four tested Babesia species was possible for each of the four species used as a reference—each species had unique difference curve shape
Agarose gel electrophoresis allowed discrimination between B. microti and the three other species: B. canis, B. divergens, B. venatorum, 408 and 381 bp fragments, respectively (Fig. 4).
Gel electrophoresis of multiplex PCR HRM products. Agarose gel electrophoresis allowed discrimination between the four tested Babesia species. The mPCR-HRM products obtained for each species were separated in 3% agarose gel in the following order: (1) B. canis, (2) B. microti, (3) B. divergens, and (4) B. venatorum. M: 100 bp DNA ladder (GPB 600 bp DNA Ladder, GenoPlast)
The mPCR-HRM detection limit of B. microti and B. divergens was at least 5 copies of 18S rRNA target sequences (Fig. 5), detected in 6 repeats, i.e., in the three samples in the two independent mPCR-HRM reactions with 100% positive results.
In silico analyses of mPCR-HRM specificity revealed the presence of perfect binding sites for the primer B-rev in at least 18 Babesia taxons and multiple perfect and imperfect binding sites for the forward primers (Table 2). Perfect binding sites for primer pairs, B-BDV/B-rev and B-BM/B-rev, were present in 5 and 2 Babesia species, respectively (Table 2). The primer pair B-BM/B-rev has perfect binding site in the genome of Theileria sp. HN1 [GenBank: FJ645726.1] (Table S2).
In silico no significant similarity was found between the designed primer pairs and the DNA of the following: human, dog, ticks, roe deer, ungulates, Plasmodium falciparum, and Toxoplasma gondii. The specificity was confirmed experimentally in mPCR-HRM with negative control DNA samples.
Ten blood samples from dogs with confirmed B. canis infection and 5 Microtus oeconomus blood samples with confirmed B. microti infection gave reproducible, positive results of mPCR-HRM species determination (Fig. S1).
Discussion
The most important feature of the designed mPCR-HRM method is the reduction of the procedure of DNA detection and identification of several Babesia species to a single tube reaction.
The clinical course of babesiosis in immunocompromised patients is often severe, and the mortality rate in such cases may reach 20% (Levin and Krause 2016). In contrast, in adult immunocompetent persons, who can be blood donors, the course of Babesia infection is asymptomatic. In most such cases, low level parasitemia persists in blood for a few months from the day of the infection (Krause et al. 1998; Vannier and Krause 2012).
There has been growing concern about the risk of B. microti transmission by blood and blood derived products in the USA (Levin and Krause 2016; Simon et al. 2014). The discussions on the necessity of blood screening to prevent transfusion transmitted babesiosis and on optimal screening tools continue (Bish et al. 2015; Leiby 2006; Levin and Krause 2016; Simon et al. 2014). However, it is known that the screening costs should not exceed its benefit for public health (Levin and Krause 2016; Simon et al. 2014).
The designed multiplex PCR HRM method is a tool for detection of infections with at least three Babesia species pathogenic to humans, in a single tube assay, which might match economic criteria for babesiosis blood screening tests. The assay was proven to have no cross reactions with protozoan parasites Toxoplasma gondii and Plasmodium falciparum related to Babesia. Such a cross reaction may pose a problem in serology.
Ideally, the sensitivity of microscopy examination, still a “gold standard” in diagnostics of intraerythrocytic protozoa, should reach a detection limit of 10–50 parasites/μl of blood in a thick blood smear in a person with a normal red blood cell count (Guerin et al. 2002; Kamau et al. 2011; Moody 2002; Wang et al. 2015b). However, most laboratories require ten-fold higher parasitemia (100–500) to detect intraerythrocytic parasite infection (development of recommendations for the protection of short-stay travelers to malaria endemic areas: Memorandum from two WHO Meetings 1988; Milne et al. 1994; Wang et al. 2015b). Furthermore, the infected red blood cell count can vary depending on the person performing microscopy examination of the same blood sample (Wang et al. 2015b).
The sensitivity of most PCR methods for B. microti detection, employing DNA isolation procedures used routinely in blood diagnostics, was estimated to be within the range 1–30 parasites per one 1 μl of blood (Chan et al. 2013; Rollend et al. 2013; Wang et al. 2015a, b). The only method claimed to have higher sensitivity (Bloch et al. 2013) used a non-standard approach for DNA isolation which is not used in routine blood examination performed in medical diagnostics (Wang et al. 2015b). Our assay detects at least 5 copies of the Babesia spp. 18S rRNA fragment in 1 μl of solution added to PCR. The sensitivity was tested using the DNA of B. microti and B. divergens (Fig. 5); thus, both primer pairs used in mPCR-HRM were tested: B-BM/B-rev and B-BDV/B-rev, respectively. At least two copies of the target 18S rRNA are present in one Babesia cell (Cornillot et al. 2012); therefore, with parasitemia of 1–30 parasites per one 1 μl of blood, at least 2–60 copies of the Babesia 18S rRNA gene should be present in each microliter of blood. Since in the DNA isolation step, 1 ml of blood is converted into 100 μl of DNA solution, there should be around 20–600 copies of target sequence per microliter of the DNA solution. Therefore, mPCR-HRM sensitivity should be at least comparable to that of other PCR methods used for Babesia detection. However, we agree with the previously expressed opinion that direct comparison of the Babesia PCR detection methods is required for proper evaluation of their sensitivity (Wang et al. 2015b).
It should be emphasized that most existing PCR methods used for diagnosis of babesiosis in humans allow detection of single species infection—B. microti (Hildebrandt et al. 2013; Wang et al. 2015b; Wang et al. 2015a; Bloch et al. 2013; Rollend et al. 2013; Chan et al. 2013). In the USA, most infections are caused by B. microti while in Europe, most infections are caused by other Babesia species (Hildebrandt et al. 2013). The demand for multiplex PCR for detection of several Babesia species from blood has been expressed, and our mPCR-HRM method might be one of the possible ways to fill the demand (Hildebrandt et al. 2013). We experimentally confirmed the ability of mPCR-HRM to detect and differentiate at least three species of Babesia infecting humans: B. microti, B. venatorum, and B. divergens. The possibility of species determination could be important not only for epidemiology or basic research but also for medicine as due to possible differences in the clinical course of the infections caused by distinct Babesia species, different approaches towards treatment may be required (Ord and Lobo 2015).
In silico analysis showed that the primer B-rev was universal for at least 18 taxons of Babesia (Babesia sp. and Babesia species-like) (Table 2). The limited species specificity was achieved by application of forward primers, B-BM B-BDV, matching perfectly at least seven Babesia taxons (Table 2) including the five described to infect humans: B. microti, B. microti-like, B. divergens, B. divergens-like, B. venatorum. The DNA region spanned by those primers is polymorphic and relatively short (Fig. 1) which allows efficient amplification and differentiation of PCR products by HRM curve analysis (Fig. 2 and Fig. 3). The ability to differentiate Babesia spp. using HRM curves of PCR products was confirmed on samples containing DNA of B. microti, B. divergens, B. venatorum, and B. canis (Fig. 2 and Fig. 3).
Based on the test results obtained for B. microti, B. divergens, and B. venatorum and in silico analysis, it seems highly probable that B. capreoli and B. odocoilei which have the perfectly matching binding sites for primers used in mPCR-HRM will be amplified and their differentiation by curve shape will be possible. To the best of our knowledge, there have been no descriptions of human infections with these species of Babesia, but they could be within the scope of interests of biologists investigating wildlife.
In general, PCR amplification may occur even if some mismatches between the primers and the DNA template are present. Based on our experience with universal primers for filarial parasite detection in mosquitoes (Masny et al. 2016), we expected that detection of other Babesia species could be possible using mPCR-HRM with the designed primers. The DNA of B. canis, species specific for dogs, was amplified in the clinical samples of infected dogs by the primer pair B-BDV, B-rev, and HRM curve analysis allowed B. canis identification (Fig. S1). The ability to amplify B. canis DNA was perceived by us as an advantage—the range of detected Babesia species was broader than we initially expected to obtain. An experiment performed on clinical blood samples of ten dogs infected with Babesia canis showed reproducible results (Fig. S1) which confirmed that the mPCR-HRM could be applicable for laboratory diagnostics of canine babesiosis and that the presence of mismatches in one primer did not have a detrimental effect on the HRM curve-based species identification reproducibility. The reproducibility of HRM curve shapes was high also for B. microti DNA isolated from naturally infected Microtus oeconomus (Fig. S1).
Organisms currently classified as Theileria spp., for example Theileria sp. HN1 (FJ645726.1), could be theoretically amplified by mPCR-HRM since it has perfect binding sites to our primers (Table S2). The classification to either Theileria or Babesia genus has been undergoing changes recently. Furthermore, classification to the same genus, Theileria or Babesia, might not reflect the real phylogenetic distance between organisms (Harris 2016). Thus, the mPCR-HRM test might be applicable to the piroplasms from both Theileria and Babesia genera which could be an advantage for researchers looking for new hosts of those parasites. Furthermore, Babesia taxons such as Babesia microti-like or Babesia divergens-like (also referred to as Babesia microti-related or Babesia cf. microti) are diverse and most probably include many separate species (Harris 2016; Baneth et al. 2015). Since the primers B-BDV, B-BM, and B-rev match sequences of multiple Babesia microti-like and Babesia divergens-like taxon representatives, it is difficult to tell how many Babesia species could be amplified with mPCR-HRM.
Screening of livestock for Babesia spp. revealed livestock infections with Babesia species not considered specific for the hosts in which those species were found. Based on the screening results, it was concluded that host specificity of Babesia was likely to be wider than it had been thought to be (Li et al. 2015). It cannot be excluded that a similar phenomenon might be observed in humans and using an assay detecting multiple Babesia species might lead to revealing piroplasm infections previously undescribed in people—new risk factors.
The combination of the ability to amplify a number of Babesia species and the low level of cross-reactivity with the host DNA (no cross-reactivity for the hosts tested here) could be desirable features in the screening studies. We have experimentally shown that mPCR-HRM was applicable to Babesia detection in different host samples, and we have not observed cross-reactivity with host DNA, i.e., tick, human, rodent, and deer. The latter is the host of B. divergens and B. venatorum infecting humans and recently is considered a likely reservoir of B. venatorum (Michel et al. 2014).
Other real-time assays using universal primers for Babesia species detection exist. For the survey of livestock pan-Babesia FRET-quantitative PCR was designed with a pair of universal primers and molecular probes capable to detect multiple species of Babesia with relatively low level of interspecies discrimination. Therefore, the final species differentiation step was sequencing (Li et al. 2015). In this respect, the pan-Babesia FRET-quantitative PCR assay is similar to other PCR assays with universal primers (Li et al. 2015; Bonnet et al. 2007). We deliberately chose HRM based identification due to the ability to differentiate known and unknown sequences and its lower costs compared to molecular probe based assays. The region spanned by the designed B-primers is polymorphic, and species-specific probes could be designed. Theoretically, the use of species-specific molecular probes might have the advantage of assuring higher specificity compared to mPCR-HRM at the price of lower range of detected parasites (with a single probe) and/or higher cost in the case of multiple Babesia species detection (multiple probes).
Amplification of Babesia DNA has been gaining popularity in detecting the parasites presence in the vectors of the disease—ticks (Venclikova et al. 2015; Orkun et al. 2014; Sytykiewicz et al. 2012; Mierzejewska et al. 2015). A strong positive association was found between the B. microti infection ratios in humans and ticks from the same territories (Diuk-Wasser et al. 2014). In our study, Babesia positive and negative tick DNA samples were selected based on the results of commonly used universal PCR with primer pair Bab-GF2 and Bab-GR2 followed by amplification with species specific primers and then by PCR product sequencing (Bonnet et al. 2007). Infected tick was detected and the species specific melting curve was obtained (manuscript in preparation). Instead of several steps (PCR, nested PCR, sequencing) (Mierzejewska et al. 2015), a single PCR HRM reaction for both detection and amplification might be applied to screening of ticks. Thus, mPCR-HRM could be a useful, time and resource saving tool for tick population screening for the assessment of the epidemiological risks associated to tick bite exposure.
The mPCR-HRM can be converted into multiplex PCR with detection of the products by gel electrophoresis. However, in such a version, the discrimination between species is limited to B. microti and the remaining ones (Fig. 4). This could be a solution for smaller laboratories where mPCR-HRM products could be sequenced only if the identification of the Babesia species was necessary. We have intended to present an inexpensive versatile tool for distinct types of screening studies; therefore, the protocols we applied to DNA isolation from various sources, i.e., blood, tissues of vertebrates, and from ticks, are similar and rely on the same set of reagents which can be prepared in-house (Boom et al. 1999; Masny et al. 2016). Apart from being cheaper, the in-house DNA isolation protocol may assure better sensitivity of the PCR detection compared to commercial kits (Schuurman et al. 2005).
To the best of our knowledge, PCR assay for detection and species identification of several Babesia species infecting humans and animals, in a single tube reaction, without application of molecular probes, has not been described to date. The range of detected Babesia species and lack of cross-reactivity with the DNA of various Babesia hosts, combined with relatively good sensitivity, might make mPCR-HRM a cost-effective tool for diagnostics and screening studies.
References
Baneth G, Florin-Christensen M, Cardoso L, Schnittger L (2015) Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasit Vectors 8:207. doi:10.1186/s13071-015-0830-5
Bish EK, Moritz ED, El-Amine H, Bish DR, Stramer SL (2015) Cost-effectiveness of Babesia microti antibody and nucleic acid blood donation screening using results from prospective investigational studies. Transfusion 55(9):2256–2271. doi:10.1111/trf.13136
Bloch EM, Lee TH, Krause PJ, Telford SR 3rd, Montalvo L, Chafets D, Usmani-Brown S, Lepore TJ, Busch MP (2013) Development of a real-time polymerase chain reaction assay for sensitive detection and quantitation of Babesia microti infection. Transfusion 53(10):2299–2306. doi:10.1111/trf.12098
Bonnet S, Jouglin M, L'Hostis M, Chauvin A (2007) Babesia sp. EU1 from roe deer and transmission within Ixodes ricinus. Emerg Infect Dis 13(8):1208–1210
Boom R, Sol C, Weel J, Gerrits Y, de Boer M, Wertheim-van Dillen P (1999) A highly sensitive assay for detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence. J Clin Microbiol 37(5):1489–1497
Canever MF, Vieira LL, Reck C, Richter L, Miletti LC (2014) First evaluation of an outbreak of bovine babesiosis and anaplasmosis in southern Brazil using multiplex PCR. Korean J Parasitol 52(5):507–511. doi:10.3347/kjp.2014.52.5.507
Chan K, Marras SA, Parveen N (2013) Sensitive multiplex PCR assay to differentiate Lyme spirochetes and emerging pathogens Anaplasma phagocytophilum and Babesia microti. BMC Microbiol 13:295. doi:10.1186/1471-2180-13-295
Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, Vacherie B, Augagneur Y, Brès V, Duclos A, Randazzo S, Carcy B, Debierre-Grockiego F, Delbecq S, Moubri-Ménage K, Shams-Eldin H, Usmani-Brown S, Bringaud F, Wincker P, Vivarès CP, Schwarz RT, Schetters TP, Krause PJ, Gorenflot A, Berry V, Barbe V, Ben Mamoun C (2012) Sequencing of the smallest apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res 40(18):9102–9114. doi:10.1093/nar/gks700
Development of recommendations for the protection of short-stay travellers to malaria endemic areas: Memorandum from two WHO Meetings (1988) Bull World Health Organ 66(2):177–196
Diuk-Wasser MA, Liu Y, Steeves TK, Folsom-O'Keefe C, Dardick KR, Lepore T, Bent SJ, Usmani-Brown S, Telford SR 3rd, Fish D, Krause PJ (2014) Monitoring human babesiosis emergence through vector surveillance New England, USA. Emerg Infect Dis 20(2):225–231. doi:10.3201/eid1302/130644
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224. doi:10.1093/molbev/msp259
Guerin PJ, Olliaro P, Nosten F, Druilhe P, Laxminarayan R, Binka F, Kilama WL, Ford N, White NJ (2002) Malaria: current status of control, diagnosis, treatment, and a proposed agenda for research and development. Lancet Infect Dis 2(9):564–573
Haapasalo K, Suomalainen P, Sukura A, Siikamaki H, Jokiranta TS (2010) Fatal babesiosis in man, Finland, 2004. Emerg Infect Dis 16(7):1116–1118. doi:10.3201/eid1607.091905
Harris DJ (2016) Naming no names: comments on the taxonomy of small piroplasmids in canids. Parasit Vectors 9(1):289. doi:10.1186/s13071-016-1567-5
Herwaldt BL, Cacciò S, Gherlinzoni F, Aspöck H, Slemenda SB, Piccaluga P, Martinelli G, Edelhofer R, Hollenstein U, Poletti G, Pampiglione S, Löschenberger K, Tura S, Pieniazek NJ (2003) Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg Infect Dis 9(8):942–948
Hildebrandt A, Gray JS, Hunfeld KP (2013) Human babesiosis in Europe: what clinicians need to know. Infection 41(6):1057–1072. doi:10.1007/s15010-013-0526-8
Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF (2011) Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J Clin Microbiol 49(8):2946–2953. doi:10.1128/JCM.00276-11
Krause PJ, Spielman A, Telford SR 3rd, Sikand VK, McKay K, Christianson D, Pollack RJ, Brassard P, Magera J, Ryan R, Persing DH (1998) Persistent parasitemia after acute babesiosis. N Engl J Med 339(3):160–165
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Leiby DA (2006) Babesiosis and blood transfusion: flying under the radar. Vox Sang 90(3):157–165
Levin AE, Krause PJ (2016) Transfusion-transmitted babesiosis: is it time to screen the blood supply? Curr Opin Hematol 23(6):573–580
Li J, Kelly P, Zhang J, Xu C, Wang C (2015) Development of a pan-Babesia FRET-qPCR and a survey of livestock from five Caribbean islands. BMC Vet Res 11:246. doi:10.1186/s12917-015-0560-0
Masny A, Lewin T, Salamatin R, Golab E (2011) The first report on detection of canine Acantocheilonema reconditum in Poland and the associated diagnostic problems. Pol J Vet Sci 14(3):485–487
Masny A, Sałamatin R, Rozej-Bielicka W, Golab E (2016) Is molecular xenomonitoring of mosquitoes for Dirofilaria repens suitable for dirofilariosis surveillance in endemic regions? Parasitol Res 115(2):511–525
Michel AO, Mathis A, Ryser-Degiorgis MP (2014) Babesia spp. in European wild ruminant species: parasite diversity and risk factors for infection. Vet Res 45:65. doi:10.1186/1297-9716-45-65
Mierzejewska EJ, Pawełczyk A, Radkowski M, Welc-Falęciak R, Bajer A (2015) Pathogens vectored by the tick, Dermacentor reticulatus, in endemic regions and zones of expansion in Poland. Parasit Vectors 8:490. doi:10.1186/s13071-015-1099-4
Milne LM, Kyi MS, Chiodini PL, Warhurst DC (1994) Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol 47(8):740–742
Moody A (2002) Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15(1):66–78
Ord RL, Lobo CA (2015) Human Babesiosis: pathogens, prevalence, diagnosis and treatment. Curr Clin Microbiol Rep 2(4):173–181
Orkun Ö, Karaer Z, Çakmak A, Nalbantoğlu S (2014) Identification of tick-borne pathogens in ticks feeding on humans in Turkey. PLoS Negl Trop Dis 8(8):e3067. doi:10.1371/journal.pntd.0003067
Prince HE, Lapé-Nixon M, Patel H, Yeh C (2010) Comparison of the Babesia duncani (WA1) IgG detection rates among clinical sera submitted to a reference laboratory for WA1 IgG testing and blood donor specimens from diverse geographic areas of the United States. Clin Vaccine Immunol 17(11):1729–1733. doi:10.1128/CVI.00256-10
Rollend L, Bent SJ, Krause PJ, Usmani-Brown S, Steeves TK, States SL, Lepore T, Ryan R, Dias F, Ben Mamoun C, Fish D, Diuk-Wasser MA (2013) Quantitative PCR for detection of Babesia microti in Ixodes scapularis ticks and in human blood. Vector Borne Zoonotic Dis 13(11):784–790. doi:10.1089/vbz.2011.0935
Sadkowska-Todys M, Baumann-Popczyk A, Wnukowska N, Popczyk B, Kucharczyk B, Gołąb E (2015) Occurrence and prevalence of selected zoonotic agents: Echinococcus multilocularis, Trichinella spiralis and hepatitis E virus (HEV) in the population of polish hunters--results of the study conducted in 2010-2012. Przegl Epidemiol 69(4):673–378 823-827
Sawczuk M, Maciejewska A, Adamska M, Skotarczak B (2005) Roe deer (Capreolus capreolus) and red deer (Cervus elaphus) as a reservoir of protozoans from Babesia and Theileria genus in north-western Poland. Wiad Parazytol 51(3):243–247
Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA (2012) Babesia: a world emerging. Infect Genet Evol 12(8):1788–1809. doi:10.1016/j.meegid.2012.07.004
Schuurman T, van Breda A, de Boer R, Kooistra-Smid M, Beld M, Savelkoul P, Boom R (2005) Reduced PCR sensitivity due to impaired DNA recovery with the MagNA pure LC total nucleic acid isolation kit. J Clin Microbiol 43(9):4616–4622
Simon MS, Leff JA, Pandya A, Cushing M, Shaz BH, Calfee DP, Schackman BR, Mushlin AI (2014) Cost-effectiveness of blood donor screening for Babesia microti in endemic regions of the United States. Transfusion 54(3 Pt 2):889–899. doi:10.1111/trf.12492
Sytykiewicz H, Karbowiak G, Werszko J, Czerniewicz P, Sprawka I, Mitrus J (2012) Molecular screening for Bartonella henselae and Borrelia burgdorferi sensu lato co-existence within Ixodes ricinus populations in central and eastern parts of Poland. Ann Agric Environ Med 19(3):451–456
Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci S (2012) A new real-time PCR assay for improved detection of the parasite Babesia microti. J Clin Microbiol 50(3):903–908. doi:10.1128/JCM.05848-11
Vannier E, Krause PJ (2012) Human babesiosis. N Engl J Med 366(25):2397–2407. doi:10.1056/NEJMra1202018
Venclikova K, Mendel J, Betasova L, Hubalek Z, Rudolf I (2015) First evidence of Babesia venatorum and Babesia capreoli in questing Ixodes ricinus ticks in the Czech Republic. Ann Agric Environ Med 22(2):212–214. doi:10.5604/12321966.1152067
Wang G, Villafuerte P, Zhuge J, Visintainer P, Wormser GP (2015a) Comparison of a quantitative PCR assay with peripheral blood smear examination for detection and quantitation of Babesia microti infection in humans. Diagn Microbiol Infect Dis 82(2):109–113. doi:10.1016/j.diagmicrobio.2015.03.010
Wang G, Wormser GP, Zhuge J, Villafuerte P, Ip D, Zeren C, Fallon JT (2015b) Utilization of a real-time PCR assay for diagnosis of Babesia microti infection in clinical practice. Ticks Tick Borne Dis 6(3):376–382. doi:10.1016/j.ttbdis.2015.03.001
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. doi:10.1093/bioinformatics/btp033
Zhou X, Li SG, Chen SB, Wang JZ, Xu B, Zhou HJ, Ge HX, Chen JH, Hu W (2013) Co-infections with Babesia microti and Plasmodium parasites along the China-Myanmar border. Infect Dis Poverty 2(1):24. doi:10.1186/2049-9957-2-24
Zhou X, Xia S, Huang JL, Tambo E, Zhuge HX, Zhou XN (2014) Human babesiosis, an emerging tick-borne disease in the People's Republic of China. Parasit Vectors 7:509. doi:10.1186/s13071-014-0509-3
Zintl A, Mulcahy G, Skerrett HE, Taylor SM, Gray JS (2003) Babesia divergens, a bovine blood parasite of veterinary and zoonotic importance. Clin Microbiol Rev 16(4):622–636
Acknowledgements
We would like to thank Bogumila Skotarczak from Department of Genetics, University of Szczecin and Grzegorz Karbowiak from Witold Stefanski Institute of Parasitology, Polish Academy of Sciences, Warsaw for their kind gifts of biological material from Caproleus caproleus and Microtus oeconomus. We would like to thank Rafal Stachurski and Katarzyna Stachurska from Antecedo veterinary clinic for providing biological material from dogs.
Author information
Authors and Affiliations
Contributions
Designed the study: WRB, AM, and EG; performed the experiments: WRB; analyzed the data: WRB, AM, and EG; wrote the paper: WRB, AM, and EG. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Obtaining the patients’ consent for use of their samples for research purposes is a standard procedure in the NIPH-NIH Diagnostics Laboratories. A single human blood sample positive for Babesia venatorum was collected during project NN404520038 financed by the Ministry of Science and Higher Education. The granted study was approved by the Bioethics Commission in NIPH-NIH (Sadkowska-Todys et al. 2015).
Competing interests
Patent application is pending PCT/IB2016/052655
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rozej-Bielicka, W., Masny, A. & Golab, E. High-resolution melting PCR assay, applicable for diagnostics and screening studies, allowing detection and differentiation of several Babesia spp. infecting humans and animals. Parasitol Res 116, 2671–2681 (2017). https://doi.org/10.1007/s00436-017-5576-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5576-x