Abstract
Purpose
NSCLC is the most common type of lung cancers. The purpose of this study is to screen cancer-related mutations in early LUAD in China through NGS technology, determine their correlation with clinical characteristics and provide basis for treatment decisions.
Methods
In this study, we performed a 583 gene panel to detect the mutational spectrum of the tumors which were collected from 98 LUAD patients. The sequencing data and clinical characteristics were analyzed.
Results
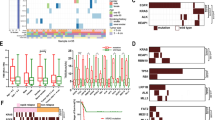
Mutations were identified in 94.9% of patients. EGFR had the highest mutation frequency which was detected in 66% of the patients and was significantly associated with female gender and non-smoking history. Other genes with high mutation frequency were TP53 (37%), ERBB2 (24%), BCOR (22%), ZFHX3 (19%), BTG1 (17%), ATR (16%), WWTR1 (15%), etc. TP53 mutations were significantly associated with medium and low differentiation of tumors; BCOR and BLM mutations with gender; WWTR1 mutations with age; and ATR mutations with visceral pleura invasion were observed. 61% of the patients harbored at less one actionable alteration associated with FDA-recognized or investigational drugs.
Conclusion
Multiple mutations in LUAD patients in this study have not previously been reported in NSCLC. Moreover, mutations in driver genes including EGFR, TP53, BCOR, BLM, WWTR1, and ATR were significantly related to clinical features. The panel used in this study is an effective approach for molecular analysis and can be applied in personalized treatment decision-making and drug development.




Similar content being viewed by others
References
Amos CI et al (1992) Host factors in lung cancer risk: a review of interdisciplinary studies. Cancer Epidemiol Biomark Prev 1:505–513
Astolfi A et al (2019) BCOR involvement in cancer. Epigenomics 11:835–855
Behnfeldt JH et al (2018) A tri-serine cluster within the topoisomerase IIalpha-interaction domain of the BLM helicase is required for regulating chromosome breakage in human cells. Hum Mol Genet 27:1241–1251
Brown NA et al (2018) Precision medicine in non-small cell lung cancer: current standards in pathology and biomarker interpretation. Am Soc Clin Oncol Educ Book 38:708–715
Cancer Genome Atlas, N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Cao Q et al (2016) BCOR regulates myeloid cell proliferation and differentiation. Leukemia 30:1155–1165
Chen WQ et al (2017) Lung cancer incidence and mortality in China in 2013. Zhonghua Zhong Liu Za Zhi 39:795–800
Chen RL et al (2018) Crizotinib in advanced non-small-cell lung cancer with concomitant ALK rearrangement and c-Met overexpression. BMC Cancer 18:1171
Diaz Z et al (2013) Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod Pathol 26:1413–1424
Duma N et al (2019) Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 94:1623–1640
Fountzilas E et al (2018) Treating EGFR-mutated oncogene-addicted advanced non-small-cell lung cancer in the era of economic crisis in Greece: challenges and opportunities. J Glob Oncol 4:1–12
Fumarola C et al (2014) Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol 90:197–207
Hensing T et al (2014) A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol 799:85–117
Hou H et al (2019) Concurrent TP53 mutations predict poor outcomes of EGFR-TKI treatments in Chinese patients with advanced NSCLC. Cancer Manag Res 11:5665–5675
Inamura K (2017) Lung cancer: understanding its molecular pathology and the 2015 WHO classification. Front Oncol 7:193
Jung KJ et al (2016) The effect of smoking on lung cancer: ethnic differences and the smoking paradox. Epidemiol Health 38:e2016060
Kwak EL et al (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363:1693–1703
Labbe C et al (2017) Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 111:23–29
Lee PN et al (2018) The relationship of cigarette smoking in Japan to lung cancer, COPD, ischemic heart disease and stroke: a systematic review. F1000Res 7:204
Lee YS et al (2014) Genomic profile analysis of diffuse-type gastric cancers. Genome Biol 15:R55
Liu Y et al (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49:1211–1218
Liu H et al (2018) Multifaceted regulation and functions of YAP/TAZ in tumors (review). Oncol Rep 40:16–28
Lv SD et al (2019) Integrative molecular characterization of Chinese prostate cancer specimens. Asian J Androl 22:162–168
Maki-Nevala S et al (2016) Hot spot mutations in Finnish non-small cell lung cancers. Lung Cancer 99:102–110
Margueron R et al (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349
Mayakonda A et al (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 28:1747–1756
Mazzoni SM et al (2014) AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett 355:1–8
Meldrum C et al (2011) Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev 32:177–195
Narita A et al (2019) Stereotactic body radiotherapy for primary non-small cell lung cancer patients with clinical T3–4N0M0 (UICC 8th edition): outcomes and patterns of failure. J Radiat Res 60:639–649
Pao W et al (2010) Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 10:760–774
Park SJ et al (2017) New targets in non-small cell lung cancer. Hematol Oncol Clin N Am 31:113–129
Planchard D et al (2018) Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 29:iv192–iv237
Planchard D et al (2015) Lung cancer in never-smokers. Eur Respir J 45:1214–1217
Rao CV et al (2017) Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis 38:2–11
Scheijen B et al (2017) Tumor suppressors BTG1 and IKZF1 cooperate during mouse leukemia development and increase relapse risk in B-cell precursor acute lymphoblastic leukemia patients. Haematologica 102:541–551
Sgambato A et al (2018) Targeted therapies in non-small cell lung cancer: a focus on ALK/ROS1 tyrosine kinase inhibitors. Expert Rev Anticancer Ther 18:71–80
Siegel RL et al (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30
Skov BG et al (2015) The prevalence of EGFR mutations in non-small cell lung cancer in an unselected Caucasian population. APMIS 123:108–115
Sparmann A et al (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6:846–856
Sun X et al (2007) Infrequent mutation of ATBF1 in human breast cancer. J Cancer Res Clin Oncol 133:103–105
Sun GG et al (2014) The expression of BTG1 is downregulated in NSCLC and possibly associated with tumor metastasis. Tumour Biol 35:2949–2957
Torre LA et al (2016) Lung cancer statistics. Adv Exp Med Biol 893:1–19
Tripathi V et al (2018) MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways. Nat Commun 9:1016
Vestergaard HH et al (2018) A systematic review of targeted agents for non-small cell lung cancer. Acta Oncol 57:176–186
Walker CJ et al (2015) Patterns of CTCF and ZFHX3 mutation and associated outcomes in endometrial cancer. J Natl Cancer Inst 107:djv249
Yang J et al (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181
Yu G et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Zanconato F et al (2016) YAP/TAZ at the roots of cancer. Cancer Cell 29:783–803
Zheng HC et al (2015) BTG1 expression correlates with pathogenesis, aggressive behaviors and prognosis of gastric cancer: a potential target for gene therapy. Oncotarget 6:19685–19705
Acknowledgements
The authors thank the medical workers and researchers from the department of thoracic surgery, pathology and laboratory of Peking Union Medical College Hospital for their excellent work in data collection, sequencing and analysis for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1.
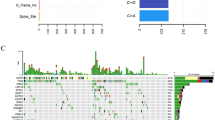
High frequency mutated genes are the driving genes of LUAD. A. Hot spot mutated sites in LUAD patients (N=98). B. Somatic interactions detected by using R. C. Plot showing tumor driver genes predicted based on positional clustering. The size of the point represents the number of clusters found in the gene. X-axis shows number of mutations in these clusters. (TIF 1333 kb)
Supplementary Figure 2.
The mutated sites of genes with high mutation frequency. (PDF 300 kb)
Rights and permissions
About this article
Cite this article
Zhou, X., Xu, X., Tian, Z. et al. Mutational profiling of lung adenocarcinoma in China detected by next-generation sequencing. J Cancer Res Clin Oncol 146, 2277–2287 (2020). https://doi.org/10.1007/s00432-020-03284-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-020-03284-w