Abstract
Introduction
HNF4α expression and SMARCA4 loss were thought to be features of non-terminal respiratory unit (TRU)-type lung adenocarcinomas, but their relationships remained unclear.
Materials and methods
HNF4α-positive cases among 241 lung adenocarcinomas were stratified based on TTF-1 and SMARCA4 expressions, histological subtypes, and driver mutations. Immunohistochemical analysis was performed using xenograft tumors of lung adenocarcinoma cell lines with high HNF4A expression.
Result
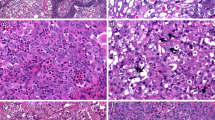
HNF4α-positive adenocarcinomas(n = 33) were divided into two groups: the variant group(15 mucinous, 2 enteric, and 1 colloid), where SMARCA4 was retained in all cases, and the conventional non-mucinous group(6 papillary, 5 solid, and 4 acinar), where SMARCA4 was lost in 3/15 cases(20%). All variant cases were negative for TTF-1 and showed wild-type EGFR and frequent KRAS mutations(10/18, 56%). The non-mucinous group was further divided into two groups: TRU-type(n = 7), which was positive for TTF-1 and showed predominantly papillary histology(6/7, 86%) and EGFR mutations(3/7, 43%), and non-TRU-type(n = 8), which was negative for TTF-1, showed frequent loss of SMARCA4(2/8, 25%) and predominantly solid histology(4/8, 50%), and never harbored EGFR mutations. Survival analysis of 230 cases based on histological grading and HNF4α expression revealed that HNF4α-positive poorly differentiated (grade 3) adenocarcinoma showed the worst prognosis. Among 39 cell lines, A549 showed the highest level of HNF4A, immunohistochemically HNF4α expression positive and SMARCA4 lost, and exhibited non-mucinous, high-grade morphology in xenograft tumors.
Conclusion
HNF4α-positive non-mucinous adenocarcinomas included TRU-type and non-TRU-type cases; the latter tended to exhibit the high-grade phenotype with frequent loss of SMARCA4, and A549 was a representative cell line.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer-related death in many developed countries, including the United States and Japan [1, 2]. Adenocarcinoma is the most common histological subtype of lung cancer [3].
The existence of a distinct subset of lung adenocarcinomas arising from the terminal respiratory unit (TRU) was previously proposed by Yatabe et al. [4,5,6]. TRU-type lung adenocarcinomas, which are estimated to account for 75%–80% of primary lung adenocarcinomas, show histologically non-mucinous lepidic growth or papillary components and frequently express thyroid transcription factor-1 (TTF-1), which is the master regulator of lung differentiation at high levels [4,5,6]. The genetic backgrounds of TRU-type adenocarcinomas have been investigated in detail. Epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) fusions were found to be specific to TRU-type adenocarcinomas [7, 8]. However, limited information is currently available on non-TRU-type lung adenocarcinomas.
Non-TRU-type lung adenocarcinomas are not a single entity but include various histological and molecular subtypes [9,10,11]. Kim et al. reported that mucinous adenocarcinomas without TTF-1 expression can be regarded as non-TRU-type lung adenocarcinomas [12], and Yatabe et al. reported that the representative non-TRU-type lung adenocarcinomas were poorly differentiated and exhibited solid morphology [4]. Our previous report revealed that the main group of non-TRU-type lung adenocarcinomas were hepatocyte nuclear factor 4α (HNF4α)-positive adenocarcinomas with gastrointestinal features that frequently harbored KRAS mutations and TTF-1 inactivating mutations/hypermethylation [11].
HNF4α is one of the ligand-dependent transcription factors and specifically expressed in the liver and gastrointestinal organs (stomach, small intestine, and pancreas) but not in normal human lung tissue [13, 14]. HNF4α regulates epithelial cell polarity and morphogenesis and plays an important role in gastrointestinal and hepatic cell differentiation [15,16,17,18]. HNF4α also has a role as an oncoprotein and is involved in carcinogenesis, cancer growth, and invasion in various cancers such as hepatocellular cancer, colorectal cancer, gastric cancer, and Barret’s esophageal cancer [19,20,21,22]. In the field of lung adenocarcinoma, HNF4α was first reported as a characteristic marker for invasive mucinous adenocarcinomas (IMA) [23], which were regarded as non-TRU-type lung adenocarcinomas. However, the frequency of HNF4α expression in adenocarcinomas other than IMA is not well recognized, especially in non-mucinous lung adenocarcinomas.
SMARCA4 is one of the catalytic subunits in SWI/SNIF chromatin remodeling complexes and has recently been suggested as a tumor suppressor [24,25,26,27,28]. We previously reported that the inactivating mutations of SMARCA4 were correlated with the epithelial-mesenchymal transition (EMT) phenotype of lung adenocarcinoma cell lines, and loss of SMARCA4 expression was frequent in poorly differentiated non-TRU-type adenocarcinomas, showing a lack of lepidic growth, low expressions of TTF-1 and wild-type EGFR [28]. Both the expression of HNF4α and the loss of SMARCA4 are considered characteristics of non-TRU-type adenocarcinomas, but their relationship remains unclear.
This is the first report focusing on the relationships among immunohistochemical expression patterns of HNF4α, TTF-1, and SMARCA4, histological subtypes, and driver mutations. The whole sections of 241 primary lung adenocarcinomas were used in this study. HNF4α expression was found not only in mucinous, enteric, and colloid adenocarcinomas but also in morphologically conventional non-mucinous adenocarcinomas. Some of them heterogeneously expressed HNF4α and TTF-1, which were mutually exclusive within the same tumor. These cases were considered TRU-type adenocarcinomas and frequently harbored EGFR mutations. Moreover, TTF-1-negative and HNF4α-positive non-mucinous adenocarcinomas showed wild-type EGFR and frequent SMARCA4 loss, and tended to show a high-grade solid morphology and very poor prognosis.
We also examined the histological and immunohistochemical features of xenograft tumors derived from lung adenocarcinoma cell lines. The HNF4α-positive lung adenocarcinoma cell lines (A549, Calu3, H1651, and H2405) all showed non-mucinous and high-grade morphology, and the A549 cell line showed a marked loss of SMARCA4, indicating that it was a representative cell line of HNF4α-positive, non-mucinous lung adenocarcinoma with high-grade morphology.
Materials and methods
Case selections
Details are shown in Online Resource 1.
Histological analysis
Details are shown in Online Resource 1.
Immunohistochemical analysis
Detailed staining and evaluation protocols are shown in Online Resource 2.
Sequencing using a next-generation sequencer
Mutations of primary lung tumors were investigated using the MINtS system, employing a MiSeq sequencer (Illumina K.K.), as previously reported [29]. The protocol of RNA extraction is shown in Online Resource 3.
Cell lines and medium
We used 39 non-squamous non-small-cell lung cancer cell lines. Detailed information is available in our previous reports [9, 28, 30,31,32,33,34].
Mutational analysis of the 39 cell lines
Gene mutations in the 39 cell lines were based on our previous reports [9, 28, 30,31,32,33,34] and data from the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle/).
Gene expression profile and single nucleotide polymorphism array analyses of 39 lung adenocarcinoma cell lines
A comprehensive gene expression analysis was undertaken using an oligonucleotide microarray (GeneChip Human Genome U133A; Affymetrix), as previously described [35,36,37]. Analysis with a single nucleotide polymorphism array (Human Mappings 50 K Xbal array; Affymetrix) was performed using the Genome Imbalance Map algorithm, as previously described [38].
Xenograft tissues of lung adenocarcinoma cell lines
Details are shown in Online resource 4.
Statistical analysis
For all statistical analyses, SPSS 26 (SPSS, Chicago, IL, USA) was used. Correlations between clinicopathological features and HNF4α expression were analyzed using the χ2 test. The Kaplan–Meier method was used for the calculation of survival curves, and the Wilcoxon method was used for comparisons. Multivariate analysis was performed using the Cox proportional hazards model. Differences were considered significant for p-values < 0.05.
Results
Clinicopathological features of HNF4α-positive adenocarcinomas
We conducted an immunohistochemical analysis of HNF4α using 241 primary lung adenocarcinoma samples surgically resected at Jichi Medical University Hospital and found that 33 samples (14%) were positive for HNF4α. Table 1 shows the relationships between HNF4α expression and the clinicopathological features of 238 patients (241 samples). A total of 6 lung adenocarcinoma samples from the 3 patients with double primary lung adenocarcinomas were all positive for TTF-1 and negative for HNF4α. All samples of mucinous (15/15, 100%), enteric (2/2, 100%), and colloid (1/1, 100%) adenocarcinoma exhibited HNF4α expression. HNF4α expression was detected in a proportion of acinar (4/24, 17%), papillary (6/123, 5%), and solid (5/43, 12%) adenocarcinomas. Representative figures of HNF4α-positive lung adenocarcinomas are shown in Fig. 1. None of the HNF4α-positive lung adenocarcinomas showed hepatoid differentiation. None of the in-situ non-mucinous, minimally invasive, or lepidic adenocarcinoma samples (WHO grade 1), representing TRU-type adenocarcinomas, exhibited HNF4α expression.
Table 1 also shows the correlations among HNF4α expression levels and driver mutations, clinicopathological factors and immunohistochemical patterns. In HNF4α-positive samples, the frequency of KRAS mutations was significantly high (20/33, 61%) (p < 0.001), and the frequency of EGFR mutations was significantly low (3/33, 9%) (p < 0.001), whereas no common drive mutations other than KRAS and EGFR (e.g., ALK, HER2, MET, BRAF, RET, or ROS1) were found. EGFR and KRAS mutations were mutually exclusive.
HNF4α expression was correlated with the advanced pT stage (pT2-pT4) (p = 0.001) and STAS (p = 0.001), but not correlated with pleural invasion, lymphatic or vessel invasion, intrapulmonary metastasis, or nodal involvement.
Immunohistochemically, HNF4α expression was correlated with a loss of SMARCA4 (p = 0.035) and MUC5AC expression (p < 0.001), and inversely correlated with the expression of TTF-1 (p < 0.001) (Table 1), but seven samples were double-positive for TTF-1 and HNF4α, including six papillary adenocarcinomas and one solid adenocarcinoma. Although the loss of SMARCA2 was not significantly more frequent in HNF4α-positive adenocarcinomas, two of the four HNF4α-positive Grade 3 adenocarcinomas that expressed SMARCA4 showed the loss of SMARCA2.
TTF-1 and SMARCA4 expression and gene mutation patterns differed in HNF4α-positive lung adenocarcinomas according to histology
Based on the 2021 WHO classification of thoracic tumors [39], we divided HNF4α-positive adenocarcinoma cases (n = 33) into two groups: the variant group (mucinous, enteric, and colloid adenocarcinomas) (n = 18) and the conventional non-mucinous group (acinar, papillary, and solid adenocarcinomas) (n = 15) (Fig. 2a). All variant group cases were diffusely HNF4α-positive and completely TTF-1-negative. None of them harbored EGFR mutations, but more than half of the cases harbored the KRAS mutation (10/18, 55.6%). In contrast, almost half of the cases in the non-mucinous group were double-positive for TTF-1 and HNF4α (7/15, 46.7%), and their expression patterns were heterogenous and mutually exclusive within the same tumor (Online Resource 5a).
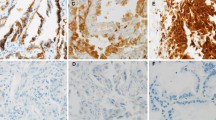
a The histological subtypes (mucinous, enteric, colloid, papillary, acinar, and solid adenocarcinomas), histological grades, immunohistochemical expression of HNF4α, TTF-1, SMARCA4, SMARCA2 and MUC5AC and genetic mutations of EGFR and KRAS in 33 HNF4α-positive lung adenocarcinoma cases, with division into the variant and non-mucinous groups. b HE, SMARCA4, HNF4α, and TTF-1 staining of representative cases of HNF4α-positive non-mucinous adenocarcinomas with loss of SMARCA4 (Cases 22 and 26). Both cases were grade 3 adenocarcinomas, SMARCA4 lost, HNF4α-positive, and TTF-1-negative. Note that lymphoid cells within the tumor were SMARCA4-positive (100 × magnification, Scale bar: 100 μm)
The three EGFR-mutated cases in the non-mucinous group were all double-positive for TTF-1 and HNF4α. Given the high frequency of EGFR mutations in these double-positive cases (3/7, 43%), we speculated that the double-positive adenocarcinomas were of the TRU-type and that TTF-1-positive TRU-type adenocarcinomas were induced to express HNF4α through the local loss of TTF-1 (e.g., by epigenetic silencing). In addition, all cases in the variant group retained SMARCA4 expression, but in the non-mucinous group, loss of SMARCA4 was detected in 3 of the 15 cases (20%), much more frequently than in HNF4α-negative non-mucinous adenocarcinomas (3/208, 1.4%) (Fig. 2a and Table 1). Figure 2b shows histological images of two representative cases of HNF4α-positive non-mucinous adenocarcinoma with the loss of SMARCA4.
The loss of SMARCA2, a paralog of SMARCA4, did not correlate with the expression of HNF4α (Table 1) and was detected among HNFα-positive cases in both the variant group (5.6%, 1/18) and conventional non-mucinous group (13.3%, 2/15) (Fig. 2a and Online Resource 5b). MUC5AC expression was frequently positive in HNF4α-positive cases (in both the variant and conventional groups), but was almost negative in TTF-1-positive cases (6/7, 85.7%) (Fig. 2a and Online Resource 5b).
HNF4α-positive non-mucinous adenocarcinomas with high-grade morphology (WHO grade 3) showed the worst prognosis
The three-tiered grading system is the common prognostic indicator of non-mucinous lung adenocarcinomas [39]. In the present study, the 5-year survival rates of grade 1 (n = 29), grade 2 (n = 128), and grade 3 (n = 56) groups were 100%, 86.0%, and 61.4% respectively, and the survival rates differed significantly (grade 1 vs. grade 2: p = 0.032, grade 2 vs. grade 3: p = 0.002) (Fig. 3a). Next, for survival analysis, we re-classified non-mucinous adenocarcinoma cases of each grade group into HNF4α-positive and HNF4α-negative groups: HNF4α-positive grade 3 group (n = 6), HNF4α-negative grade 3 group (n = 50), HNF4α-positive grade 2 group (n = 9), HNF4α-negative grade 2 group (n = 119), and HNF4α-negative grade 1 group (n = 29), as well as the variant group (n = 17). Notably, the HNF4α-positive grade 3 group showed worse prognosis than the HNF4α-negative grade 3 group (3-year survival rates of 51.4% and 69.3%, respectively) (p = 0.024), showing the worst prognosis among the six groups (Fig. 3b).
a Overall survival among 213 cases of non-mucinous adenocarcinomas categorized according to the WHO grading system. b The prognoses of 230 lung adenocarcinomas were analyzed in 6 groups; HNF4α + G3: HNF4α-positive grade 3 (n = 6), HNF4α-G3: HNF4α-negative grade 3 (n = 50), HNF4α + G2: HNF4α-positive grade 2 (n = 9), HNF4α-G2: HNF4α-negative grade 2 (n = 119), HNF4α-G1:HNF4α-negative grade 1 (n = 29), and the variant group (n = 17). The samples with unknown prognoses (n = 5) and double carcinoma cases (n = 3) were excluded
We found that in grade 3 non-mucinous adenocarcinomas (n = 56), sex, pleural invasion, pStage, HNF4α expression and MUC5AC expressions, were poor prognostic factors (Online Resource 6a). We performed a multivariate analysis, excluding the expression of MUC5AC, which correlated with the expression of HNF4α, and found that the expression of HNF4α and the pStage remained significant in the multivariate analysis (HR, 3.318; CI, 1.344–8.188 for HNF4α expression and HR, 9.019; CI, 4.107–19.804 for pStage) (Online Resource 6b). Although HNF4α-positive grade 3 non-mucinous adenocarcinomas frequently showed the loss of SMARCA4 (2/6, 33%), it was not identified as a poor prognostic factor (Online Resource 6a).
We also compared clinicopathological factors among the six groups (Online Resource 7) and found that advanced pT factor, advanced pStage, lymph node metastasis, vessel invasion, pleural invasion, and pulmonary metastasis were most frequently observed in the HNF4α-positive grade 3 group, indicating that this group was the aggressive phenotype.
Xenograft tumors of HNF4α-positive lung adenocarcinoma cell lines showed high-grade, non-mucinous morphology
Finally, we examined whether HNF4α-positive grade 3 adenocarcinoma cell lines were present among the 39 non-squamous non-small cell lung cancer cell lines. Online Resource 8 shows the gene-level expressions of HNF4A and TTF-1 in the 39 cell lines. The four cell lines with the highest expression of HNF4A were A549, H2405, Calu-3, and H1651, in that order. Online Resource 8 also shows the common driver mutations of the 39 cell lines, and among the four HNF4A-high cell lines, SMARCA4 and KRAS mutations were found in A549, HER2 amplification was found in Calu-3, and no common driver mutations were found in H2405 or H1651.
Figure 4a summarizes (i) the genetic status of EGFR, MET, HER2, KRAS, and SMARCA4 (upper panel), (ii) gene-level expressions of HNF4A, TTF-1, and SMARCA4 (middle panel), and (iii) protein-level expressions of HNF4α, TTF-1, SMARCA4, and ACTB (lower panel) for the four HNF4A-high cell lines (A549, H2405, H1651, and Calu3), compared with the four representative TRU-type cell lines with TTF-1-high expressions (HCC827, PC3, H1648, and H2009). The four HNF4A-high cell lines showed high HNF4α expression and low level of TTF-1, except for H1651, at both the gene and protein levels. A marked decrease in the expression level of SMARCA4 was only observed in SMARCA4-mutated A549, whereas the other three HNF4A-high cell lines exhibited SMARCA4 expression. An aberrant band of SMARCA4 was detected in H2405 by western blot analysis (Fig. 4a).
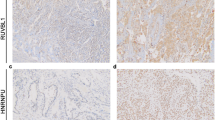
a Genetic status of EGFR, MET, HER2, KRAS, and SMARCA4 (upper panel), gene-level expressions of HNF4A, TTF-1, and SMARCA4 (middle panel), and protein expression levels of HNF4α, TTF-1, SMARCA4, and ACTB (lower panel) for 8 cell lines, including the four cell lines that highly express HNF4A (A549, H2405, H1651, and Calu-3) and the four cell lines that highly express TTF-1 (HCC827, PC3, H1648, and H2009). In the upper panel, the gray box indicates the presence of genetic abnormalities and the white box indicates the absence of genetic abnormalities. In the middle lane, red means more than or equal to the average of each gene expression, orange means under the average but more than or equal to one-quarter of the average, and green means under one-quarter of the average. b The histological features and immunohistochemical expression patterns of HNF4α, TTF-1, and SMARCA4 for the xenograft tumors of A549, H2405, H1651, and Calu-3
Next, using xenograft tumors of the four HNF4A-high cell lines, we examined the histological growth patterns in HE staining and performed immunohistochemical analysis for HNF4α, TTF-1, and SMARCA4 (Fig. 4b). A549 and H1651 showed solid growth patterns, H2405 showed solid growth patterns with focal cribriform patterns, and Calu-3 showed fused glandular and papillary growth patterns (Fig. 4b, the top row). All of these growth patterns are features of grade 3 primary lung adenocarcinoma, and notably, none of the cell lines showed morphological features of mucinous adenocarcinoma. Immunohistochemically, all of the four HNF4A-high cell lines were HNF4α-positive and TTF-1-negative in the nucleus (Fig. 4b, the second and third row), but H1651, which showed high TTF-1 expression at both the gene and protein levels, exhibited intracytoplasmic TTF-1 expression. SMARCA4 expression was diffusely lost in the A549 xenograft tumor but retained in the other three cell lines (Fig. 4b, the bottom row).
Discussion
Here, we have shown that HNF4α expression was not limited to mucinous, enteric, or colloid adenocarcinomas, which showed gastrointestinal morphology, but also appeared in morphologically conventional non-mucinous adenocarcinomas such as acinar, papillary, and solid adenocarcinomas.
In the present study (mostly Asian cases), the frequency of KRAS mutations was significantly higher in HNF4α-positive adenocarcinomas (39.4%, 13/33 cases) than in HNF4α-negative adenocarcinomas (9.2%, 19/207 cases). Based on The Cancer Genome Atlas data of 456 primary lung adenocarcinomas (mostly Caucasian cases), the frequency of KRAS mutations was not significantly higher in HNF4A-high cases (35.9%, 42/117 cases) than in HNF4A-low cases (29.5%, 100/339 cases) (p = 0.121) (Online resource 9). KRAS mutations in lung adenocarcinoma are more frequent in Caucasians than in Asians. We speculate that this is not because of the higher frequency of HNF4α-positive cases in Caucasians, but because of the higher frequency of KRAS mutations in HNF4α-negative adenocarcinomas (mainly TRU-type lung adenocarcinomas) in Caucasians.
The results obtained herein revealed the absence of common driver mutations other than KRAS mutations in mucinous adenocarcinomas (60%, 9/15 cases). NRG1 gene fusion, which has been reported in KRAS wild-type mucinous adenocarcinomas, was not examined in the present study [40]. CD74-NRG1 fusion genes have been identified as driver oncogenes and ERBB2/ERBB3 receptors may be the target of these fusion genes [41]. We previously noted that the knockdown of HNF4A in lung adenocarcinoma cell lines suppressed the expression and phosphorylation of ERBB3 (data not shown). The relationship among ERBB3, HNF4A, and NRG1 fusion genes is a topic for future studies. Although they were not identified in the present study, ALK fusion genes have been detected in mucinous adenocarcinomas [42], but are TTF-1-positive TRU-type adenocarcinomas with a mucinous morphology, a distinct entity from HNF4α-positive mucinous lung adenocarcinomas, exhibiting gastrointestinal features.
In the present study, the frequency of TTF-1 expression, loss of SMARCA4, and EGFR mutations differed according to histology. All cases of mucinous, enteric, and colloid adenocarcinoma were HNF4α-positive, TTF-1-negative, and SMARCA4 retained, and showed a high frequency of KRAS mutations (10/18, 55.6%) and no EGFR mutations (0/18, 0%). A recent study showed that 16 cases of enteric and mucinous adenocarcinoma lacked common driver mutations except for KRAS mutations [43], indicating that mucinous, enteric, and colloid adenocarcinomas might form a single spectrum of HNF4α-positive non-TRU-type adenocarcinomas showing gastrointestinal differentiation. However, enteric adenocarcinomas occasionally show focal TTF-1 expression and EGFR mutations [44], suggesting that some enteric adenocarcinomas may be phenotypically altered from TRU-type adenocarcinomas. The etiology of enteric adenocarcinoma is controversial and requires further investigation.
Approximately half of the HNF4α-positive non-mucinous adenocarcinomas were TTF-1-positive (7/15, 47%), and they frequently showing papillary predominant histology (6/7, 86%). They were often accompanied by a non-mucinous lepidic component (5/7, 71%) and often lacked MUC5AC expression (1/7, 14.3%). Furthermore, approximately half of the cases harbored EGFR mutations (3/7, 43%), suggesting that the double-positive cases for HNF4α and TTF-1 were derived from TRU-type lung adenocarcinomas. In these cases, HNF4α and TTF-1 were expressed heterogeneously and were mutually exclusive within the same tumor. We speculate that focal loss of TTF-1 expression may be partially due to TTF-1 gene hypermethylation, as previously reported [11].
The remaining half of the HNF4α-positive non-mucinous adenocarcinomas were totally TTF-1-negative (8/8, 100%) and never harbored EGFR mutations (0/8, 0%), suggesting that they were the non-TRU-type lung adenocarcinomas. Half of them were poorly differentiated solid adenocarcinomas (4/8, 50%), potentially differing from mucinous, enteric, and colloid adenocarcinomas, which showed histologically gastrointestinal differentiation. We previously reported that HNF4α was not a significant prognostic factor in lung adenocarcinomas at any stage [10], but confirmed that the expression of HNF4α and a solid morphology were independent poor prognostic factors in advanced stage samples. In this study, HNF4α-positive poorly differentiated (grade 3) non-mucinous adenocarcinomas were aggressive phenotypes and showed the worst prognosis, and HNF4α expression was an independent prognostic factor in grade 3 non-mucinous lung adenocarcinomas. These results suggest that the expression of HNF4α plays a distinctive role in the progression of lung adenocarcinoma and a poor prognosis.
HNF4α was recently shown to be involved in the growth and invasion of various cancers as oncoprotein [19,20,21,22]. A previous study that examined the expression of HNF4α and mucin profiles in lung mucinous adenocarcinomas [45] reported that HNF4α induced the expression of MUC3 in KRAS-mutated mucinous adenocarcinomas, which is a poor prognostic factor for mucinous adenocarcinomas of the breast and appendix [46, 47]. Chen et al. demonstrated that the HNF4α-BC200-FMR-positive feedback loop promoted cell growth and metastasis in KRAS-mutated, HNF4α-positive cell lines (A549) [48]. Therefore, the expression of HNF4α has potential as a therapeutic target in lung adenocarcinomas, particularly KRAS-mutated lung adenocarcinomas.
Herein, we found that HNF4α-positive non-mucinous adenocarcinomas frequently showed loss of SMARCA4 (3/15, 20%), much more frequently than in mucinous, enteric, and colloid adenocarcinomas (0/18, 0%), and HNF4α-negative non-mucinous adenocarcinomas (3/208, 1.4%). Additionally, loss of SMARCA4 was more frequently observed in HNF4α-positive grade 3 adenocarcinomas (2/6, 33%). The function of SMARCA4 varies among different organs and diseases, and SMARCA4 inactivation in lung cancer is related to the loss of lung lineage transcription and early metastasis [49]. We speculated the HNF4α expression in grade 3 adenocarcinoma may imply dedifferentiation associated with the inactivated SMARCA4 function, resulting in high-grade morphology and poor prognosis.
In the present study, loss of SMARCA4 was only found in 2.5% (6/241) of the lung adenocarcinoma samples. SMARCA4 mutation rates were reported to account for approximately 8% of non-small cell lung cancers, but not all mutations resulted in loss of SMARCA4 expression [49]. Some variants of SMARCA4 mutation may show intact SMARCA4 expression despite the loss of its function [50]. Note that, unlike HNF4α, loss of SMARCA4 was not an independent prognostic factor in grade 3 adenocarcinomas in our study (Online Resource 6), but we did not investigate the mutational status of SMARCA4 in SMARCA4-retained adenocarcinomas. Further studies are needed to elucidate the relationship between HNF4α expression and the function of SMARCA4.
We also demonstrated that four lung adenocarcinoma cell lines (A549, H1651, H2405, and Calu-3) had high HNF4α expression at both the gene and protein levels. All four cell lines tended to show high expression levels of Vimentin and ZEB1 compared with the TTF-1-high cell lines, and relatively low expression of CDH1 (Online Resource 10), indicating dedifferentiation or EMT. All four cell lines may be regarded as representatives of non-mucinous HNF4α-positive lung adenocarcinomas with grade 3 morphology, but their immunohistochemical and genetic features varied. We propose that A549 is not a mucinous adenocarcinoma cell line [48], but A549 may be a representative cell line of HNF4α-positive grade 3 lung adenocarcinomas with aggressive pathological features.
In conclusion, a subset of HNF4α-positive adenocarcinomas, such as mucinous adenocarcinomas with gastrointestinal differentiation, are TTF-1-negative and SMARCA4 retained, often showing KRAS mutations. In addition, some conventional non-mucinous adenocarcinomas are HNF4α-positive, which include not only TRU-type adenocarcinomas that are double-positive for TTF-1 and HNF4α but also non-TRU-type poorly differentiated (grade 3) adenocarcinomas with frequent loss of SMARCA4 expression. HNF4α-positive grade 3 adenocarcinoma shows a very poor prognosis, and HNF4α expression is an independent prognostic factor in grade 3 lung adenocarcinomas. Thus, examining the status of HNF4α expression is important for not only assuming the etiology and gene mutational status but also predicting the prognosis in non-mucinous adenocarcinomas. The A549 cell line may be considered a representative cell line of HNF4α-positive grade 3 adenocarcinomas.
Data availability
Raw data can be obtained from the corresponding author upon reasonable request.
Code availability
N/A.
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Katanoda K, Hori M, Saito E et al (2021) Updated trends in cancer in Japan: incidence in 1985–2015 and mortality in 1958–2018-A sign of decrease in cancer incidence. J Epidermiol 31(7):426–450. https://doi.org/10.2188/jea.JE20200416
Barta JA, Powell CA, Wisnivesky JP (2019) Global epidemiology of lung cancer. Ann Glob Health 85(1):8. https://doi.org/10.5334/aogh.2419
Yatabe Y, Mitsudomi T, Takahashi T (2002) TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol 26(6):767–773. https://doi.org/10.1097/00000478-200206000-00010
Yatabe Y, Mitsudomi T (2007) Epidermal growth factor receptor mutations in lung cancers. Pathol Int 57(5):233–244. https://doi.org/10.1111/j.1440-1827.2007.02098.x
Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T (2005) EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol 29(5):633–639. https://doi.org/10.1097/01.pas.0000157935.28066.35
Yoshida A, Tsuta K, Nakamura H et al (2005) Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol 35(8):1226–1234. https://doi.org/10.1097/PAS.0b013e3182233e06
Takeuchi K, Soda M, Togashi Y et al (2012) RET, ROS1 and ALK fusions in lung cancer. Nat Med 18(3):378–381. https://doi.org/10.1038/nm.2658
Matsubara D, Ishikawa S, Oguni S, Aburatani H, Fukayama M, Niki T (2010) Co-activation of epidermal growth factor receptor and c-MET defines a distinct subset of lung adenocarcinomas. Am J Pathol 177(5):2191–2204. https://doi.org/10.2353/ajpath.2010.100217
Matsubara D, Yoshimoto T, Soda M et al (2020) Reciprocal expression of trefoil factor-1 and thyroid transcription factor-1 in lung adenocarcinomas. Cancer sci 111(6):2183–2195. https://doi.org/10.1111/cas.14403
Matsubara D, Soda M, Yoshimoto T et al (2017) Inactivating mutations and hypermethylation of the NKX2-1/TTF-1 gene in non-terminal respiratory unit-type lung adenocarcinomas. Cancer Sci 108(9):1888–1896. https://doi.org/10.1111/cas.13313
Kim YK, Shin DH, Kim KB et al (2015) MUC5AC and MUC5B enhance the characterization of mucinous adenocarcinomas of the lung and predict poor prognosis. Histopathology 67(4):520–528. https://doi.org/10.1111/his.12693
Yeh MM, Bosch DE, Daoud SS (2019) Role of hepatocyte nuclear factor 4-alpha in gastrointestinal and liver diseases. World J of Gastroenterol 25(30):4074–4091. https://doi.org/10.3748/wjg.v25.i30.4074
Tanaka T, Jiang S, Hotta H et al (2006) Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol 208(5):662–672. https://doi.org/10.1002/path.1928
Babeu JP, Darsigny M, Lussier CR, Boudreau F (2009) Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol 297(1):G124-134. https://doi.org/10.1152/ajpgi.90690.2008
Tuncer S, Sade-Memisoglu A, Keskus AG et al (2020) Enhanced expression of HNF4α during intestinal epithelial differentiation is involved in the activation of ER stress. FEBS J 287(12):2504–2523. https://doi.org/10.1111/febs.15152
Moore BD, Khurana SS, Huh WJ, Mills JC (2016) Hepatocyte nuclear factor 4α is required for cell differentiation and homeostasis in the adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol 311(2):G267–G275. https://doi.org/10.1152/ajpgi.00195.2016
Li J, Nign G, Duncan SA (2000) Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev 14(4):464–474
Chang HR, Nam S, Kook MC et al (2016) HNF4α is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut 65(1):19–32. https://doi.org/10.1136/gutjnl-2014-307918
Huang Y, Xian L, Liu Z et al (2022) AMPKα2/HNF4A/BORIS/GLUT4 pathway promotes hepatocellular carcinoma cell invasion and metastasis in low glucose microinviroment. Biochem Pharmacol 203:115198. https://doi.org/10.1016/j.bcp.2022.115198
Darsigny M, Babeu JP, Seidman EG et al (2010) Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res 70(22):9423–9433. https://doi.org/10.1158/0008-5472.CAN-10-1697
Nowicki-Osuch K, Zhuang L, Jammula S et al (2021) Molecular phenotyping reveals the identity of Barrett’s esophagus and its malignant transition. Science 373(6556):760–767. https://doi.org/10.1126/science.abd1449
Sugano M, Nagasaka T, Sasaki E et al (2013) HNF4α as a marker for invasive mucinous adenocarcinoma of the lung. Am J Surg Pathol 37(2):211–218. https://doi.org/10.1097/PAS.0b013e31826be303
Medina PP, Romero OA, Kohno T et al (2008) Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat 29(5):617–622. https://doi.org/10.1002/humu.20730
Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D (2008) Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res 68(10):3689–3696. https://doi.org/10.1158/0008-5472.CAN-07-6652
Romero OA, Setien F, John S et al (2012) The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med 4(7):603–616. https://doi.org/10.1002/emmm.201200236
Fukuoka J, Fujii T, Shih JH et al (2004) Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res 10(13):4314–4324. https://doi.org/10.1158/1078-0432.CCR-03-0489
Matsubara D, Kishaba Y, Ishikawa S et al (2013) Lung cancer with loss of BRG1/BRM, shows epithelial mesenchymal transition phenotype and distinct histologic and genetic features. Cancer Sci 104(2):266–273. https://doi.org/10.1111/cas.12065
Inoue Y, Shiihara J, Miyazawa H et al (2017) A highly specific and sensitive massive parallel sequencer-based test for somatic mutations in non-small cell lung cancer. PLoS ONE 12(4):e0176525. https://doi.org/10.1371/journal.pone.0176525
Matsubara D, Kanai Y, Ishikawa S et al (2012) Identification of CCDC6-RET fusion in the human lung adenocarcinoma cell line, LC-2/ad. J Thorac Oncol 7(12):1872–1876. https://doi.org/10.1097/JTO.0b013e3182721ed1
Matsubara D, Ishikawa S, Oguni S, Aburatani H, Fulayama M, Niki T (2010) Molecular predictors of sensitivity to the MET inhibitor PHA665752 in lung carcinoma cells. J Thorac Oncol 5(9):1317–1324. https://doi.org/10.1097/JTO.0b013e3181e2a409
Ito T, Matsubara D, Tanaka I et al (2016) Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci 107(10):1527–1538. https://doi.org/10.1111/cas.13013
Matsubara D, Yoshimoto T, Akolekar N et al (2023) Genetic and phenotypic determinants of morphologies in 3D cultures and xenografts of lung tumor cell lines. Cancer Sci 114(4):1757–1770. https://doi.org/10.1111/cas.15702
Ibrahim R, Matsubara D, Osman W et al (2014) Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol 45(7):1397–1405. https://doi.org/10.1016/j.humpath.2014.02.013
Ishii M, Hashimoto S, Tsutsumi S et al (2000) Direct comparison of GeneChip and SAGE on the quantitative accuracy in transcript profiling analysis. Genomics 68(2):136–143. https://doi.org/10.1006/geno.2000.6284
Midorikawa Y, Yamamoto S, Ishikawa S et al (2006) Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene 25(40):5581–5590. https://doi.org/10.1038/sj.onc.1209537
Wang T, Niki T, Goto A et al (2007) Hypoxia increases the motility of lung adenocarcinoma cell line A549 via activation of the epidermal growth factor receptor pathway. Cancer sci 98(4):506–511. https://doi.org/10.1111/j.1349-7006.2007.00428.x
Ishikawa S, Komura D, Tsuji S et al (2005) Allelic dosage analysis with genotyping microarrays. Biochem Biophys Res Commun 333(4):1309–1314. https://doi.org/10.1016/j.bbrc.2005.06.040
WHO Classification of Tumours Editorial Board (2021) Thoracic Tumours. Thoracic Tumours. 5th ed. Lyon, France: International Agency for Research on Cancer
Shim HS, Kenudson M, Zheng Z et al (2015) Unique Genetic and Survival Characteristics of Invasive Mucinous Adenocarcinoma of the Lung. J Thorac Oncol 10(8):1156–1162. https://doi.org/10.1097/JTO.0000000000000579
Werr L, Plenker D, Dammert MA et al (2022) CD74-NRG1 Fusions Are Oncogenic In Vivo and Induce Therapeutically Tractable ERBB2:ERBB3 Heterodimerization. Mol Cancer Ther 21(5):821–830. https://doi.org/10.1158/1535-7163
Duruisseaux M, Antoine M, Rabbe N et al (2017) Lepidic predominant adenocarcinoma and invasive mucinous adenocarcinoma of the lung exhibit specific mucin expression in relation with oncogenic drivers. Lung Cancer 109:92–100. https://doi.org/10.1016/j.lungcan.2017.05.007
Palmirotta R, Lovero D, D’Oronzo S et al (2020) Pulmonary enteric adenocarcinoma: an overview. Expert Rev Mol Med 22:e1. https://doi.org/10.1017/erm.2020.2
Okada F, Takeda M, Fujii T et al (2024) Clinicopathological and genetic analyses of pulmonary enteric adenocarcinoma. J Clin Pathol 77(2):111–115. https://doi.org/10.1136/jcp-2022-208583
Guo M, Tomoshige K, Meister M et al (2017) Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Mol Med 9(4):462–481. https://doi.org/10.15252/emmm.201606711
Rakha EA, Boyce RW, Abd El-Rehim D et al (2005) Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol 18(10):1295–1304. https://doi.org/10.1038/modpathol.3800445
Shibahara H, Higashi M, Yokoyama S et al (2014) A comprehensive expression analysis of mucins in appendiceal carcinoma in a multicenter study: MUC3 is a novel prognostic factor. PLoS ONE 9(12):e115613. https://doi.org/10.1371/journal.pone.0115613
Chen X, Zhao Y, Wang D et al (2021) The HNF4α-BC200-FMR1–Positive Feedback Loop Promotes Growth and Metastasis in Invasive Mucinous Lung Adenocarcinoma. Cancer Res 81(23):5904–5918. https://doi.org/10.1158/0008-5472.CAN-21-0980
Concepcion CP, Ma S, LaFave LM et al (2022) Smarca4 inactivation promotes lineage-specific transformation and early metastatic features in the lung. Cancer Disdov 12(2):562–585. https://doi.org/10.1158/2159-8290.CD-21-0248
Schoenfeld AJ, Bandlamudi C, Lavery JA et al (2020) The genomic landscape of SMARCA4 alterations and associations with outcomes in patients with lung cancer. Clin Cancer Res 26(21):5701–5708. https://doi.org/10.1158/1078-0432.CCR-20-1825
Funding
The present study was supported in part by JSPS KAKENHI (Grant Nos. 23K14469 to Hitomi Kawai, and 19K07441 and 22H02840 to Daisuke Matsubara, 18H02634 to Toshihiro Niki and 17K15652 to Taichiro Yoshimoto), the Foundation for Development of the Community, and Ministry of Education, Culture, Sports, Science and Technology (MEXT)-Supported Programs for the Strategic Research Foundation at Private Universities (2013–2019 and 2016–2021).
Author information
Authors and Affiliations
Contributions
Conceptualization: Hitomi Kawai, Tamaki Miura, and Natsumi Kawamatsu; Methodology: Tomoki Nakagawa, Aya Shiba-Ishii, Taichiro Yoshimoto, Yusuke Amano, and Atsuhi Kihara; Formal analysis and investigation: Yuji Sakuma, Kazutaka Fujita, Tomoki Shibani, and Shumpei Ishikawa; Writing—original draft preparation: Hitomi Kawai and Tamaki Miura; Writing—review and editing: Tetsuo Ashiko, Masashi Fukayama, and Daisuke Matsubara; Funding acquisition: Hitomi Kawai; Resources: Hisayoshi Tsubochi, Shunsuke Endo, and Koichi Hagiwara; Supervision: Daisuke Matsubara and Toshiro Niki.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the Institutional Ethics Review Committee at Jichi Medical University, Tochigi, Japan.
Financial or non-financial interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kawai, H., Miura, T., Kawamatsu, N. et al. Expression patterns of HNF4α, TTF-1, and SMARCA4 in lung adenocarcinomas: impacts on clinicopathological and genetic features. Virchows Arch (2024). https://doi.org/10.1007/s00428-024-03816-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00428-024-03816-6