Abstract
Main conclusion
In this review, we summarize how chlorophyll metabolism in angiosperm is affected by the environmental factors: light, temperature, metal ions, water, oxygen, and altitude.
Abstract
The significance of chlorophyll (Chl) in plant leaf morphogenesis and photosynthesis cannot be overstated. Over time, researchers have made significant advancements in comprehending the biosynthetic pathway of Chl in angiosperms, along with the pivotal enzymes and genes involved in this process, particularly those related to heme synthesis and light-responsive mechanisms. Various environmental factors influence the stability of Chl content in angiosperms by modulating Chl metabolic pathways. Understanding the interplay between plants Chl metabolism and environmental factors has been a prominent research topic. This review mainly focuses on angiosperms, provides an overview of the regulatory mechanisms governing Chl metabolism, and the impact of environmental factors such as light, temperature, metal ions (iron and magnesium), water, oxygen, and altitude on Chl metabolism. Understanding these effects is crucial for comprehending and preserving the homeostasis of Chl metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
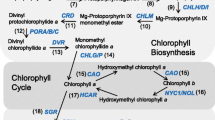
Chlorophyll (Chl) is the predominant pigment in the biosphere and plays a crucial role in the photosynthesis of angiosperms. The attainment of dynamic stability in Chl content relies on accurate and efficient Chl metabolism. The mechanisms underlying Chl synthesis and degradation during angiosperms development have been extensively investigated for several decades (Tanaka and Tanaka 2006; Wang et al. 2020; Wang and Grimm 2021). Previous studies on Chl metabolism have been extensively investigated, thus we will provide a concise overview here (Fig. 1). Chl synthesis is dependent on the tetrapyrrole biosynthesis (TPB) pathway (Brzezowski et al. 2015; Tanaka and Tanaka 2007; Vavilin and Vermaas 2002). In Arabidopsis (Arabidopsis thaliana), the initial step involves the rate-limiting enzyme glutamate 1-semialdehyde aminotransferase (GSA-AT) and glutamyl-tRNA reductase (GluTR), which convert glutamyl-tRNAGlu into 5-aminolevulinic acid (ALA). A series of subsequent transformations converts ALA into protoporphyrin IX (Proto IX), a common precursor of Chl and heme. Proto IX becomes Magnesium-Proto IX (Mg-Proto IX) with Mg2+ in the presence of Mg-chelatase (MgCh), and Mg-Proto IX will lose Mg2+ to form protochlorophyllide (Pchlide). Subsequently, the light-dependent Pchlide oxidoreductase (LPOR, hereinafter referred to as POR) (Reinbothe et al. 2010; Nguyen et al. 2021) works with divinyl reductase (DVR) (Wang and Grimm 2021) to produce chlorophyllide (Chlide) a. Chl synthase (CHLG) then catalyzes the formation of the hydrophobic Chl a by integrating the phytyl pyrophosphate into Chlide a (Lin et al. 2014). Chl a can be further reversibly converted to Chl b by “Chl cycle”, and these newly synthesized Chl a and Chl b rapidly integrate into the Chl-binding proteins (CBPs) of the photosystem (PS)-light-harvesting complexes (LHC) (Wang and Grimm 2021). The first crucial enzyme in the Chl degradation pathway is MgCh [STAYGREEN/NON-YELLOWING (SGR/NYE)], as removal of the core Mg2+ from Chl a irreversibly leads to its destruction (Chen et al. 2016). During leaf senescence, Chl a dissociates from the pigment-protein complex and undergoes breakdown through the pheophorbide a oxygenase (PAO)/phyllobilin pathway. The conversion of Chl b to Chl a is essential for the Chl degradation, as only Chl a and phosphorous pheophorbide are accepted as substrates by both NYEs/SGRs and PAO (Kruse et al. 1995; Qian et al. 2016). The primary fluorescent Chl catabolite (pFCC) and hydroxy-pFCC, generated through a series of reactions, are transported from the chloroplast and subsequently enter the vacuole after modification. Plants do not accumulate red Chl catabolites (RCCs), which are rapidly reduced to pFCCs. Deficiency in PAO or RCCR can accelerate cell death due to the accumulation of substrates for these enzymes (Pružinská et al. 2003). The Chl metabolic pathway in plants is highly intricate, involving various enzymes and genes, playing an indispensable role in the normal growth and development of angiosperms.
A schematic model of the chlorophyll (Chl) metabolic pathway, represented by Arabidopsis thaliana. Arrows indicate reactions. The enzymes involved are listed next to the arrows. The genes involved are in the blue boxes. Black dots represent Mg2+ and red dots represent Fe2+. Yellow prisms represent Proto IX. Light green prisms with black dot and yellow prism with red dot represent Mg-Proto IX and Heme, respectively. Green prisms with black dot represent Chlorophyllide a and Chl a, and dark green prisms with black dot represent Chlorophyllide b and Chl b
Since angiosperms are unable to migrate, the environment has a significant influence on their Chl metabolism. Although Chl metabolism has some commonalities in other organisms, this review focuses on the intricate interplay between angiosperms physiology and environmental factors, including light, temperature, metal ions, water, oxygen, and altitude in order to maintain a delicate balance in Chl metabolism.
Chlorophyll metabolism is dependent on Chl synthesis and degradation genes
Chl metabolism is dependent on the expression of numerous Chl biosynthesis genes (CBGs) and Chl catabolism genes (CCGs), and the enzymes and major genes involved in each important reaction step are enumerated in Fig. 1. The stay-green trait is observed in plants lacking Chl b reductase (CBR) (Sato et al. 2009), 7-hydroxymethyl Chl a reductase (HCAR) (Meguro et al. 2011), Mg dechelatase (SGR/NYE) (Chen et al. 2016) or pheophytinase (PPH) (Schelbert et al. 2009). In contrast to other Chl catabolic enzymes (CCEs), overexpression of SGR/NYE isoforms leads premature yellowing of leaves by promoting Chl breakdown in young leaves (Sakuraba et al. 2014b; Wu et al. 2016; Shimoda et al. 2016). The C-terminal cysteine-rich motif (Xie et al. 2019) of SGR/NYE1 is indispensable for its conserved role in Chl degradation (Qian et al. 2016). Studies have demonstrated that mutations in homologs of SGR1/NYE1 lead to the retention of a green phenotype in various plant species, including Arabidopsis (Ren et al. 2007), rice (Park et al. 2007), pea (Sato et al. 2007), tomato (Barry et al. 2008), and brassica napus (Qian et al. 2016). SGR contributes to the deterioration of the photosystem in addition to Chl degradation (Shimoda et al. 2016).
Chlorophyll biosynthesis requires a balance of chlorophyll and heme
In the TPB pathway that occurs in plastids, a crucial heme branch exists where Mg (Mg2+) ions are inserted into Proto IX to generate Chl, while ferrous (Fe2+) ions are inserted into Proto IX to generate heme. The initiation of Chl production relies on MgCh, an ATP-dependent heteromeric polymerase composed of the catalytic H subunit (CHLH) and the two AAA+ proteins CHLD and CHLI (Axelsson et al. 2006). Upon the binding of the H subunit to the substrate Proto IX in complex with ATP-I-D-Mg2+, allosteric changes are induced in the D subunit, thereby activating the ATPase activity of the I subunit for ATP hydrolysis, which supplies energy for Mg chelation. Subsequently, this leads to the formation of a holoenzyme by chelating Mg2+ into Proto IX, resulting in the completion of the entire catalytic process. The CHLI mutation resulted in yellow-green strawberry leaves and reduced Chl levels. Under high light intensity (300 μmol m−2 s−1), the mutant exhibited impaired chloroplast development, decreased photosynthetic capacity, significantly reduced Mg-Proto IX, and impaired Chl biosynthesis. Interestingly, this mutant demonstrated a certain tolerance to low light (50 μmol m−2 s−1), with normal chloroplast development, increased Chl content, and higher photosynthetic capacity compared to the wild type (Ma et al. 2023). This suggests a potential effect of light on MgCh enzyme activity. Heterozygous barley mutants with deficiencies in MgCh also displayed decreased Chl biosynthesis (Braumann et al. 2014).
The conversion of Proto IX to heme is catalyzed by ferrochelatase (FC), and subsequently, heme is transformed into phytochromobilin (Tanaka and Tanaka 2007). Inhibition of FC1 expression results in a reduction in heme levels. However, this does not affect Chl levels or the performance of photosystem II (PSII) (Nagai et al. 2007). A high-Chl mutant dg was identified in a library of Chinese cabbage mutants, which maintains a fine-tuned balance between heme and Chl synthesis through a regulatory mechanism that enhances Chl synthesis via dBrFC2-mediated promotion of BrPORB enzymatic responses. An amino acid mutation in the Chl a/b-binding motif (CAB) of ferrochelatase 2 (BrFC2) in dg was observed to enhance the formation of BrFC2 homodimers, thereby promoting heme production. Both wild-type BrFC2 and mutant dBrFC2 interacted with Pchlide oxidoreductases B1 and B2 (BrPORB1 and BrPORB2), but dBrFC2 exhibited a higher affinity for the substrate Pchlide, consequently promoting an elevation in Chl content (Liu et al. 2022). The regulation of heme and Chl synthesis during TPB plays a crucial role in maintaining optimal Chl levels, ultimately enhancing photosynthetic efficiency.
Post-translational coordination of chlorophyll synthesis and breakdown
The majority of research on Chl metabolism has primarily focused on transcriptional regulation. However, recent studies have made significant advancements in understanding post-translational processes. Several post-translational factors have been identified to regulate TPB, including GluTR-Binding protein (GBP) (Czarnecki et al. 2011), the chaperone Chloroplast Signal Recognition Particle 43 (cpSRP43) (Wang et al. 2018b), and LHC-LIKE 3 (Tanaka et al. 2010). GluTR is considered a pivotal regulatory enzyme that undergoes tight control at both transcriptional and post-translational levels. GBP (previously known as PROTON GRADIENT REGULATION7), a constituent of the thylakoid membrane's 300-kD protein complex, facilitates heme synthesis by retaining a fraction of GluTR on the plastid membrane responsible for ALA transportation into the heme biosynthesis pathway (Czarnecki et al. 2011). cpSRP43 mediates targeting and insertion of light-harvesting Chl a/b-binding proteins (LHCPs) and directly interacts with GluTR, indicating a crucial mechanism for post-translational coordination between LHCP insertion and Chl biosynthesis. Moreover, cpSRP43 and GBP exert distinct effects on the stability of GluTR (Wang et al. 2018b). It has been discovered that cpSRP43 interact with PORB, safeguarding its stability and normal functionality to ensure efficient Chl synthesis during leaf greening and heat shock responses (Ji et al. 2023). Although several post-translational control mechanisms regulating the Chl biosynthesis pathway have been characterized (Czarnecki and Grimm 2012; Brzezowski et al. 2015), further investigations are warranted to comprehensively elucidate the post-translational regulation of Chl catabolic metabolism, encompassing the activity, stability, and suborganelle localization of diverse genes associated with Chl metabolism (Hörtensteiner and Kräutler 2011; Kuai et al. 2018).
The precise regulation of Chl levels is crucial for optimizing photosynthesis and plant adaptation, which relies on maintaining a balance between Chl anabolism and catabolism rates (Fig. 2). GENOMES UNCOUPLED 4 (GUN4) has emerged as a key post-translational regulator of Chl biosynthesis (Peter and Grimm 2009). GUN4 plays a role in plastidic retrograde signaling, binds to Proto IX and Mg-Proto IX, and activates MgCh (Larkin et al. 2003). Previously, it was believed that GUN4 enhances the interactions between CHLH and chloroplast membranes, which serve as the site of MgCh activity, by forming more stable connections with porphyrins (Adhikari et al. 2011). Overexpression of GUN4 frequently results in the activation of enzymes, involved in Chl biosynthesis, whereas deficiency of GUN4 hampers ALA synthesis and Chl accumulation (Peter and Grimm 2009). BALANCE OF Chl METABOLISM 1 and 2 (BCM1 and BCM2) regulate post-translational dynamics of Chl during leaf development (Wang et al. 2020). BCM1, an intrinsic membrane protein, plays conserved roles in both Chl metabolic pathways and is predominantly expressed during seedling growth. In response to light, BCM1 interacts with GUN4 to enhance MgCh activity and promote Chl biosynthesis (Zhang et al. 2020; Wang et al. 2020). Conversely, the expression of BCM2 is upregulated during leaf senescence. BCMs are highly conserved regulators of Chl metabolism in plants, with their direct homologs being selected during the domestication of the tomato, soybean, and rice (Luquez and Guiamét 2001; Wang et al. 2018a). Studies have demonstrated that both BCMs function as negative regulators of Chl catabolism by interacting with SGR1/NYE1. Overexpression of either BCM1 or BCM2 delays Chl degradation and maintains a green phenotype, while inactivation of both BCMs result in a premature aging phenotype. Importantly, it should be noted that alterations in Arabidopsis' levels of BCM do not impact the expression of CBGs or CCGs (Wang et al. 2020).
Post-translational coordination of Chl synthesis and breakdown in Arabidopsis leaves is regulated by GUN4. GUN4 interacts with CHLH, binds to the intermediates Proto IX and Mg-Proto IX, and activates Mg-chelatase (MgCh, consisting of the subunits CHLH, CHLI, and CHLD) as a post-translational regulator of Chl metabolism (Adhikari et al. 2011; Zhang et al. 2021). Early in the development of the leaf, BCM1 promotes MgCh through the action of GUN4, interacts with Mg-protoporphyrin IX methyltransferase (CHLM), thus promoting Chl biosynthesis, and interacts with SGR1, destabilizing SGR1 and preventing Chl decomposition (Wang et al. 2020; Yamatani et al. 2022). Newly synthesized Chl rapidly binds to various Chl-binding proteins in the photosystem (PS)-light-harvesting complex (LHC). BCM1 is severely suppressed during the start of leaf senescence, while BCM2 and SGR1 are upregulated. Although BCM2 prevents Chl catabolism, the large accumulation of SGR1 eventually leads to Chl catabolism in plants
Imbalance of chlorophyll metabolism and the presence of antioxidant mechanisms
Reactive oxygen species (ROS) can be excessively generated in plants under abiotic or biotic stress, resulting in cellular damage (oxidative stress) and even cell death in severe cases (Apel and Hirt 2004). ROS are frequently produced because of various metabolic pathways in plants, including the Chl metabolic pathway, and ROS in turn destroy Chl molecules and/or Chl precursor (Chakraborty and Tripathy 1992; Santabarbara et al. 2002; Pattanayak and Tripathy 2011). The effects of light (e.g., high irradiance, fluctuating light, and short wavelengths of light) may damage photosynthetic proteins, leading to a decrease in photosynthetic efficiency and the formation of photodamage. The imbalance between PSII photodamage and its subsequent repair leads to photoinhibition. To prevent the accumulation of photodamaged PSII, photosynthetic organisms need to undergo repair, which involves the destruction and regeneration of D1 protein, and inhibit the repair of PSII by inhibiting the synthesis of PSII proteins (Takahashi and Murata 2008). Generally, rapid integration of Chl supply into CBPs occurs to prevent accumulation of phototoxic Chl and its metabolic intermediates (Allen et al. 2011). Given that both wavelength and photosynthetic photon flux density vary throughout the day, Chl biosynthesis predominantly takes place under low light conditions, while midday and early afternoon witness PSII photoinhibition and Chl degradation. Singlet oxygen (1O2) and superoxide (O2·−) are generated in chloroplasts by PSII and PSI, respectively, under conditions of excessive light stress (Asada 2006; Waszczak et al. 2018; Smirnoff and Arnaud 2019). Dogra and Kim (2020) focused on elucidating the mechanisms underlying 1O2 production and its impact on plant physiology, providing a comprehensive account of its generation and functional role within chloroplasts. Multiple potential sites for O2·− generation in chloroplasts, including PSI, PSII, and the electron transport chain, have been identified; moreover, O2·− can give rise to H2O2. A thorough review of H2O2 metabolism within chloroplasts is available in Smirnoff and Arnaud (2019). To avoid excessive accumulation of ROS in chloroplasts, Lutein and zeaxanthin, two carotenoids present in the LHC, regulate Chl's dynamic stability by quenching 3Chl* and preventing anomalous energy transfer to surrounding molecules. However, insufficient quenching of 3Chl* can lead to its reaction with molecular oxygen (3O2) released by the water-splitting event in the oxygen-producing complex (OEC), resulting in the formation of 1O2 (van Mieghem et al. 1995; Rinalducci et al. 2004). Under mild stress conditions, a fraction of 1O2 can evade the quenching mechanism in PSII and diffuse into the thylakoid membrane, resulting in lipid peroxidation. To counteract this process, tocopherol and plastoquinone (PQ), both present in the thylakoid membrane, act as preventive measures by detoxifying 1O2 (Santabarbara et al. 2007; Li et al. 2009). Additionally, in the stroma, ascorbate can eliminate released 1O2 from the thylakoid membrane (Kruk and Trebst 2008). Plastochromanol-8 (PC-8), an efficient scavenger of 1O2, efficiently safeguards lipids against photooxidative damage in Arabidopsis plants (Szymańska and Kruk 2010; Rastogi et al. 2014). Metabolomics research has further demonstrated that three flavonoid metabolites exclusively found in late-maturing soybean seeds (farrerol-7-O-glucoside, cyanidin-3-o-(6′-o-feruloyl) glucoside, and kaempferide-3-o-(6′-malonyl) glucoside) can effectively mitigate Chl degradation by scavenging oxygen free radicals within the chloroplasts (Wang et al. 2022a). However, there is still a need to explore additional mechanisms involved in maintaining equilibrium between Chl and ROS.
Light: a key environmental factor for chlorophyll balance
Light plays a significant role in plant growth and development as both a developmental switch and an information source, modulating multiple crucial events involved in plant development (de Wit et al. 2016). Light also exerts an impact on Chl metabolism in plants, resulting primarily in variations of leaf color. A study conducted on 14 gold leaf ornamental plants revealed that the variations in ambient light intensity significantly influenced the color of gold leaf plant leaves, resulting in an observed increase in Chl content and the Chl/carotenoid ratio as the leaf position decreased (Yuan et al. 2010). Light-induced de-etiolation triggers chloroplast differentiation, and the presence of intact chloroplast structure is a prerequisite for Chl metabolism. A comprehensive review of etiolation and de-etiolation studies has been previously conducted (Armarego-Marriott et al. 2020). While chloroplasts efficiently capture light energy and autonomously adapt to changes in light conditions through diverse biochemical mechanisms (Kovács et al. 2006; Johnson et al. 2011; Nath et al. 2013; Wolf et al. 2020), photoreceptors including phytochromes (Phys), cryptochromes (Crys), ultraviolet-B resistance 8 (UVR8), along with other regulatory proteins regulating plant transcriptomic responses, perceive alterations in both light intensity and quality (Liscum et al. 2020; Cheng et al. 2021). Arabidopsis possesses at least five classes of wavelength-specific photoreceptors that perceive distinct light signals: Phys (phyA-phyE) detect red and far-red light, Crys (CRY1 and CRY2), phototropins (PHOT1 and PHOT2), as well as members of the ZEITLUPE family (ZTL, FKF1, and LKP2) sense ultraviolet (UV)-A and blue light; while UV-B RESISTANCE LOCUS 8 (UVR8) absorbs UV-B radiation (Paik and Huq 2019; Podolec et al. 2021; Cheng et al. 2021). The coordination of multiple photoreceptors and their internal signaling pathways in plants has been summarized, highlighting their regulation of various downstream responses at the molecular and physiological levels to enable adaptation and survival in a changing environment (Paik and Huq 2019).
Similar to other eukaryotes, plant gene expression is regulated at multiple levels, including chromosomal organization, transcriptional control, post-transcriptional modifications, and translational processes (Cazzonelli et al. 2020). At the transcriptional level, light-induced regulation of plant genes plays a pivotal role through various cis-acting elements such as G-box (Ezer et al. 2017; Suekawa et al. 2018), E-box (Liu et al. 2015), D-box (Guo et al. 2016) and F-box (Lee et al. 2017). Numerous crucial transcription factors involved in light-related events, such as ELONGATED HYPOCOTYL5 (HY5) (Bae and Choi 2008), PHYTOCHROME-INTERACTING FACTORs (PIFs) (Leivar and Quail 2011), and GOLDEN2-LIKE (GLK) (Powell et al. 2012; Wang et al. 2013), regulate chloroplast biogenesis and photomorphogenesis. HY5, upon exposure to UV-B radiation and visible light, binds to its gene promoter facilitating gene expression (Abbas et al. 2014; Binkert et al. 2014), while being downstream regulated by GUN5 and Heat-shock protein HSP90 for Chl biosynthesis regulation (Kindgren et al. 2012). PIFs directly bind to the promoter of the GLK1 gene under dark conditions leading to the inhibition of CBG expression. It has been demonstrated that PIF4/5 can activate CCG expression during dark-induced leaf senescence either directly or indirectly (Sakuraba et al. 2014a; Song et al. 2014). Conversely, PIF proteins undergo rapid degradation in response to light, while activated GLK1 and GLK2 transcription factors positively regulate the expression of photosynthesis-related nuclear genes and HEMA1, CUN4, CHLH, and CAO, thereby promoting Chl biosynthesis (Waters et al. 2009; Martín et al. 2016). In angiosperms, the metabolic flow of Chl biosynthesis is regulated at the step of ALA synthesis. A negative feedback loop mediated by heme and Pchlide at the GluTR level reduces 5-ALA synthesis in darkness. In Arabidopsis, SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), a trans-regulator of PPH, inhibits dark-induced oxidative stress and leaf senescence (Chen et al. 2017b). These findings collectively demonstrate that both light and darkness exert direct or indirect regulatory effects on Chl metabolism.
The TPB pathway involves a crucial light-dependent enzyme known as POR, which catalyzes the conversion of Pchlide to Chlide upon exposure to light (Gabruk and Mysliwa-Kurdziel 2015). The atomic structure of POR assemblies and its coordination with photosynthetic membrane biogenesis and Chl synthesis in plants have been reported (Nguyen et al. 2021). In the absence of light, POR filaments directly induce high-curvature tubules in the membrane with spectral properties resembling prelaminar bodies, whose light-induced decomposition provides lipids essential for thylakoid assembly (Nguyen et al. 2021). In Arabidopsis, three genes, namely PORA, PORB, and PORC, encode for the enzyme POR. PORA is expressed during the initial stage of greening, while PORB primarily maintains Chl levels throughout angiosperm development (Masuda et al. 2003). PORC expression levels were found to decrease in the dark and increase in the light (Wang et al. 2022b). The expression of POR displays distinct patterns in barley (Apel 1981) and cucumber (Fusada et al. 2000), implying the potential for diverse light regulation of these genes.
A point of interest is that light-built hormone networks have important effects on Chl metabolism. Modifications in hormone metabolism and transport mediate various light responses in plants (Kami et al. 2010). The intricate relationship between light perception and hormonal regulation in key life events has been extensively discussed (de Wit et al. 2016). Specifically, there is a growing understanding of the impact of light-induced hormone networks on Chl metabolism (Fig. 3). Research has delved into the PIF-auxin pathway in Arabidopsis seedlings, which is triggered by the deactivation of phytochrome B (phyB) due to a low red to far-red light ratio (R:FR) (Fernández-Milmanda and Ballaré 2021). PIFs play a pivotal role in repressing Chl production under dark conditions, acting as a central signaling hub that integrates light, hormonal signals, and various developmental cues (Leivar and Quail 2011). PIFs are indispensable for the negative regulation of genes involved in TPB pathway responsible for Chl synthesis. The accumulation of dark-accumulating gibberellins (GAs) contributes to PIF activation by degrading DELLA proteins (DELLAs) and relieving their inhibitory effects (Cheminant et al. 2011). Despite their ability to inhibit PIF activity through direct protein-protein interactions, DELLAs independently regulate the expression of PORA and PORB (Feng et al. 2008; de Lucas et al. 2008). In the absence of light, nuclear accumulation of PIFs suppresses photomorphogenic reactions, including Chl biosynthesis. Key regulators in ethylene (Eth) and abscisic acid (ABA) signaling pathways such as EIN3, ABI5, and ENHANCED EM LEVEL (EEL), are directly activated by PIF4 and PIF5 proteins. These aforementioned PIF proteins have previously been implicated in dark-induced leaf senescence (Sakuraba et al. 2014a; Song et al. 2014; Seaton et al. 2015; Liebsch and Keech 2016). EIN3/EIL1 and PIF1 directly bind to the promoters of POR genes, thereby activating their expression and regulating several TPB genes to prevent the accumulation of Pchlide, a phototoxic intermediate in Chl biosynthesis. By eliciting EIN3/EIL1 activation through its established signal transduction cascade, Eth enhances seedling greening (Zhong et al. 2009). In Arabidopsis cytokinin (CK) receptor mutants, the light-induced upregulation of HEMA1, CHLH, and CHL27 was attenuated. However, treatment with CK in dark-grown wild-type seedlings promoted the expression of these genes (Hedtke et al. 2012; Kobayashi et al. 2014). CK and strigolactone (SL) have been shown to elevate HY5 protein levels while inhibiting COP1 activity (Vandenbussche et al. 2007; Tsuchiya et al. 2010). The pivotal role of plant hormones in governing growth and developmental processes is indisputable; however, further investigations are warranted to elucidate the intricate interplay between environmental conditions and hormonal regulation.
Effect of the hormone network, influenced by light, on Chl metabolism. Many important tetrapyrrole biosynthesis (TPB) genes, including CHLH, GUN4, and CAO, are upregulated as a result of the photosensitive and cryptochrome (Cry) photoreceptor families (de Wit et al. 2016; Kobayashi and Masuda 2016). Under light, Phy induces phosphorylation, ubiquitination, and subsequent degradation of PHYTOCHROME-INTERACTING FACTORs (PIFs), leading to the disinhibition of the TPB genes. By destroying DELLA proteins (DELLAs) and freeing PIFs from DELLA inhibition, accumulating gibberellin (GA) in the dark aids in the activation of PIFs (Cheminant et al. 2011; de Wit et al. 2016). The EIN proteins are stabilized by ethylene (Eth) and COP1 signals, and then the PORA and PORB are upregulated by binding to their promoter region (Zhong et al. 2009). HY5 protein is degraded in the dark by COP1-mediated ubiquitination(Xu 2020). At the same time, light mediated the accumulation of HY5 protein by blocking COP 1 activity and induced the expression of GLKs, which together upregulated key TPB genes. In addition, cytokinin (CK), strigolactone (SL). and lactones inhibited COP 1 activity and increased HY5 protein levels(Vandenbussche et al. 2007; Kobayashi et al. 2014; Zhang et al. 2024). Positive and negative regulation is depicted by arrows and bars, respectively
Temperature: effects of high and low temperatures on chlorophyll metabolism
The significance of environmental temperature for plants is self-evident. On one hand, the impact of repeated and more frequent high temperatures in tropical regions on plant growth and development has raised growing concerns (Li et al. 2023; Zhu et al. 2023). High temperature accelerates Chl breakdown by upregulating genes associated with Chl degradation. Studies conducted on cucumbers have demonstrated that elevated temperatures induce the expression of ABI5 and MYB44, thereby leading to Chl degradation. ABI5 directly binds to the promoters of PPH and PAO, thereby enhancing their expression and accelerating Chl degradation. The interaction between ABI5 and MYB44 relieves the transcriptional repression of PPH and PAO, leading to a reduction in MYB44’s binding to the promoters of PPH and PAO, ultimately resulting in MYB44 degradation (Liu et al. 2023a). On the contrary, low temperatures impose a significant environmental constraint that restricts plant development and crop productivity (Khan et al. 2017; Ding and Yang 2022). Low-temperature stress adversely affects photosynthetic capacity and efficiency by influencing gas exchange, pigment content, and chloroplast growth, resulting in reduced Chl fluorescence (Farooq et al. 2009). In postharvest tea leaves, cold-induced withering inhibits Chl breakdown through a substantial reduction in the activity and gene expression of Mg-dechelatase, Chlase, and pheophorbide an oxygenase enzymes (Yu et al. 2019).
Similar to light conditions, temperature conditions are intricately associated with the formation and development of chloroplasts, which serve as the site for Chl synthesis (Schwenkert et al. 2022). Both high and low-temperature stresses have detrimental effects on normal chloroplast formation, thereby impacting Chl metabolism. A genetic module known as TT3.1–TT3.2 has been identified as a regulator of heat sensitivity, ensuring protein quality in chloroplasts and promoting heat tolerance (Zhang et al. 2022). In recent years, more attention has been given to the effect of temperature on Chl breakdown rather than its regulation of Chl synthesis, which is often linked to light signals. The temperature response, including thermomorphogenesis, is also governed by phytochrome; specifically PhyB acts as a thermal sensor that regulates the temperature response (Casal and Balasubramanian 2019). The COP1-HY5 regulatory center has been demonstrated to play a crucial role in enhancing plant freezing tolerance, as well as accurately regulating plant responses to diurnal changes. Cold-induced translocation of COP1 from the nucleus to the cytoplasm results in an accumulation of HY5, which activates CBF1 and subsequently triggers the activation of multiple cold-responsive genes (Catalá et al. 2011). It is worth noting that light and temperature, two external environmental factors, often act synergistically as signals for regulating plant growth and development. Furthermore, their impacts on Chl metabolism frequently intersect and mutually influence each other.
Metal ions: stress on chlorophyll metabolism by deficiencies of Fe and Mg
Approximately, 80% of leaf iron content is localized within chloroplasts, and plants cultivated in iron-deficient environments exhibit symptoms of chlorosis (Nam et al. 2021), with genes associated with photosynthesis and tetrapyrrole metabolism being extensively downregulated (Rodríguez-Celma et al. 2013). The development of Fe deficiency-induced chlorosis is contingent upon the effectiveness of phosphorus. Iron limitation leads to a downregulation of photosynthetic genes in a P-dependent manner, including PHT4, which encodes the chloroplast ascorbate transporter protein, and bZIP58, which encodes a nuclear transcription factor. These genes prevent the downregulation of photosynthesis genes and result in a stay-green phenotype under Fe–phosphorus deficiency (Nam et al. 2021). Optimal concentrations of iron fertilizers have the potential to restore functionality to the photosynthetic electron transport chain, enhance electron transfer efficiency, and facilitate balanced energy allocation (Gao et al. 2022). It is noteworthy that iron–sulfur [Fe–S] clusters in the photosynthetic electron chain can act as cofactors for some of the enzymes of Chl metabolism (Hauenstein et al. 2016; Lu 2018; Schmidt et al. 2020). Doubtlessly, Fe is an important metal in plant growth related to chloroplast function and efficient photosynthesis, and its importance along with manganese and copper has been reviewed in detail by Schmidt et al.(2020). Furthermore, the distribution of potassium and calcium ions in ilr3-4 leaves was also affected (Akmakjian et al. 2021). Recently, it has been reported that the calcium-dependent protein kinase CPK21/23 promotes iron uptake in plants by activating the iron transporter protein IRT1 during low iron stress conditions (Wang et al. 2023). Red light triggers the activation of calcium-dependent protein kinase CPKs via PhyB, leading to an increase in cytoplasmic calcium concentration. The red light-induced calcium signal is precisely transduced into the activation of intracellular light response genes by CPKs, which interact with red light-activated PhyB and phosphorylate it in the nucleus (Zhao et al. 2023). However, whether calcium signaling affects Chl metabolism through iron remains unknown.
As the central atom of Chl, Mg plays an important role, and replacing Mg with metals such as copper or cadmium has a toxic effect on Chl (Sarkar et al. 1994; Grajek et al. 2020). Mg2+ signals the adenylate (ATP+ADP+AMP) state of the cell, and most enzymes requiring ATP use the chelated form of ATP (MgATP) (Voon et al. 2018). Mg in chloroplasts is 20% of total Mg, but may increase to 50% under Mg-deficient or low-light conditions (Kleczkowski and Igamberdiev 2021). Insufficient Mg levels result in Chl degradation, characterized by intervein greening. Transcript levels of the gene encoding Chl a/b-binding protein 2 (CAB2) were downregulated within just 3 days after Mg deficiency was induced (Ogura et al. 2020). Comparative analysis of transcriptional and translational differences under early Mg deficiency revealed that mutants of the transcription factor HY5, H+/CATION EXCHANGER 1 and 3 (CAX1 and CAX3), and UBIQUITIN 11 (UBQ11) exhibited an earlier onset of yellowing phenotype under Mg deficiency (Li et al. 2021).The cbd1 mutant exhibited reduced Chl content and lower Mg levels in the thylakoid compared to the wild type. Additionally, both cbd1 and cbd1gun5 mutants showed an accumulation of Mg-Proto IX. CBD1/BCM1 was shown to regulate chloroplast Mg homeostasis. CBD1/BCM1 was shown to regulate chloroplast Mg homeostasis. CBD1 was verified to have Mg transport activity using a Salmonella mutant strain MM281, which lacks the Mg2+ transport system (Zhang et al. 2020). However, further exploration is needed to understand how other Chl metabolism-related genes respond to Mg stress, particularly transcription factors and genes encoding Mg transporter proteins.
Water content: drought stress causes damage to chlorophyll metabolism
Droughts, floods and other disasters can exert stress on plant growth. To address these challenges, a 5G breeding approach (Genome assembly, Germplasm characterization, Gene function identification, Genomic breeding, and Gene editing) has been proposed (Varshney et al. 2020). Transcriptome analysis of two broomcorn millet varieties with differing drought tolerance revealed downregulation of MgCh subunit D (ChlD) expression under drought stress which resulted in decreased efficiency of Mg-protoporphyrin IX production. In addition, enzymes related to Chl a and Chl b, such as CHLG and CAO, were downregulated. Conversely, the majority of genes involved in Chl degradation, including SGR, PAO, RCCR, and NOL, exhibited significant upregulation (Yuan et al. 2022). WRKY transcription factors play crucial roles in drought tolerance by regulating ROS scavenging enzymes, promoting xylem vessel development and stimulating cellulose and lignin synthesis in roots (Cai et al. 2014; Chen et al. 2017a; Zhao et al. 2018; Geng et al. 2018). In apple, MdWRKY17 has been demonstrated to effectively maintain optimal Chl levels and photosynthetic activity during drought stress by indirectly downregulating MdCLH, MdPAO, and MdRCCR while directly activating the transcription of MdSUFB, a crucial protein involved in Fe–S cluster assembly, Chl metabolism, and photosynthesis (Couturier et al. 2013; Shan et al. 2021). The phosphorylation of MdWRKY17 by the water-deficit-activated cascade of MdMEK2-MdMPK6 has been identified as indispensable for regulating the expression of MdSUFB to sustain Chl levels under mild water-deficit stress, as supported by knockdown experiments (Shan et al. 2021). Applying alpha lipoic acid, a cofactor for pyruvate dehydrogenase and glycine decarboxylase, which are specific mitochondrial enzyme complexes, to maize seedlings experiencing drought stress enhanced their Chl content and PSII activity. In alpha lipoic acid-treated seedlings under stress, the relative expression levels of the RCA and Mg-CHLI genes were significantly elevated, while the relative expression of the Chlase genes was significantly reduced compared to untreated seedlings (Sezgin et al. 2019). These findings suggest that drought stress can expedite leaf senescence and facilitate Chl degradation in various plant species. However, further investigations are warranted to explore strategies aimed at mitigating this phenomenon.
Oxygen and altitude: an oxygen-sensing mechanism for angiosperm adaptation to altitude
Adaptation of individual plants (and populations) to survive at high altitudes is crucial, and it is argued that altitude adaptation involves direct perception of oxygen concentration across different altitudinal ranges. Studies have demonstrated that the regulation of hypoxia-related genes and the steady-state level of Pchlide, a biochemical intermediate in Chl biosynthesis, are controlled by an oxygen-sensing system in response to the local environmental absolute O2 concentration among various species along different altitudes (Abbas et al. 2022). In wild populations of Arabidopsis, Solanum habrochaites, Solanum cheesmaniae, and Brachypodium distachyon growing at various altitudes from sea level to over 3,000 m, a positive correlation between Pchlide accumulation and altitude is observed. Stable Pchlide levels play a crucial role in regulating Chl synthesis, and FLU has been demonstrated to impact Chl supply under light conditions (Hou et al. 2019). The modulation of Pchlide synthesis by O2 occurs through ErfVII-mediated GluTR inactivating complex components, specifically targeting the negative regulator FLU, via the PLANT CYSTEINE OXIDASE (PCO) N-degron pathway. This adaptive mechanism enables plants to respond to local atmospheric O2 levels and potentially prevent compromising singlet oxygen ROS production associated with light exposure. Furthermore, it has been observed that materials cultivated at higher altitudes and in an oxygen concentration of 15% exhibit reduced transcript levels of PORA, PORB, and CHLM genes compared to those grown under 21% oxygen conditions (excluding CHL27 or HEMA1) (Abbas et al. 2022). These findings emphasize the significant effects of altitude and oxygen levels on the expression of genes related to Chl metabolism, but the effects of oxygen partial pressure and UV radiation levels, which change with altitude, require further discussion. As mentioned earlier, imbalances in Chl metabolism are often associated with ROS, and perhaps the decrease in oxygen partial pressure reduces the photo-oxidative bleaching of Chl pigments. However, there is still much to uncover regarding transcriptional regulation in response to environmental cues.
Conclusion
For angiosperms, Chl metabolism is not free from environmental influences, and here we summarize the topics of this review (Fig. 4). Light plays a crucial role in angiosperm morphogenesis and is the environmental factor that has received the most extensive attention in the field of Chl metabolism research. Although much progress has been made at the transcriptional level, more inputs are needed on how light affects Chl metabolism processes in networks with other important conditions (such as hormones, as described earlier) (Kami et al. 2010; Gabruk and Mysliwa-Kurdziel 2015; de Wit et al. 2016; Liu et al. 2017). In addition, the effect of alternating light and dark on protein localization and conformational changes still has great research potential. It is also interesting to note that temperature, together with light, is a network of signaling mechanisms that regulate plant growth and development, typically PhyB can be used as a thermal sensor to control temperature response (Casal and Balasubramanian 2019). Both Fe and Mg play indispensable roles in plant Chl metabolism (Li et al. 2021; Nam et al. 2021; Liu et al. 2023b). We believe that BCM1, which has only recently been revealed, appears to be closely related to Mg2+ (Zhang et al. 2020), but the upstream transcription factors on which it relies are not yet conclusive. Drought stress can expedite leaf senescence and enhance Chl degradation in a diverse range of plant species (Maroco et al. 2002; Shan et al. 2021; Yuan et al. 2022), and considering epigenetic influences on this process may contribute to a breakthrough in understanding. Furthermore, altitude and oxygen levels significantly impact the expression of genes associated with Chl metabolism (Abbas et al. 2022), but this environmental condition has been less well studied and the complexity of the environmental impacts can never be ignored. In the future, a better understanding of the complex interactions between Chl metabolism and environmental factors in angiosperms (and even in the biosphere as a whole) will be crucial for researchers to further explore and modify plants to improve yield and stress resistance.
A schematic model outlining the homeostasis of Chl metabolism with environmental factors. PhyB senses red light and represses PIFs. PIFs in the dark inhibits GLK1, which directly or indirectly regulates CCGs. PhyB degrades PIFs in the light, and the activation of downstream target genes by HY5 is able to regulate Chl synthesis. PhyB plays a key role in the regulation of low-temperature signaling, while PIF4 is important for signaling in warmer environments. Mg2+ and Fe2+ participate in Chl synthesis as important elements in the heme branch. Water, oxygen, and altitude, can affect the expression of CBGs and CCGs
Data availability
Not applicable.
Abbreviations
- Glu-tRNAGlu :
-
l-Glutamyl-tRNAGlu
- ALA:
-
5-Aminolaevulinic acid
- ALAD:
-
5-Aminolaevulinic acid dehydratase
- CAO:
-
Chlide a oxygenase
- CBGs:
-
Chl biosynthesis genes
- CBPs:
-
Chl-binding proteins
- CBR:
-
Chl b reductase
- CCEs:
-
Chl catabolic enzymes
- CCGs:
-
Chl catabolism genes
- CHLG:
-
Chl synthase
- Chlide:
-
Chlorophyllide
- CLD:
-
Chl dephytylase
- CPO:
-
Coproporphyrinogen III oxidase
- Crys:
-
Cryptochromes
- DVR:
-
Divinyl reductase
- GBP:
-
GluTR-binding protein
- GluTR:
-
Glutamyl-tRNA reductase
- GSA-AT:
-
Glutamate 1-semialdehyde aminotransferase
- HCAR:
-
7-Hydroxymethyl Chl a reductase
- HMBS:
-
Hydroxymethybilane synthase
- LHC:
-
Light-harvesting complexs
- LHCPs:
-
Light-harvesting Chl a/b-binding proteins
- LPOR:
-
Light-dependent protochlorophyllide oxidoreductase
- MgCh:
-
Mg-chelatase
- MgPCY:
-
Mg-protoporphyrin IX monomethylester cyclase
- MgPMT:
-
Mg-protoporphyrin IX methyltransferase
- Mg-Proto IX:
-
Magnesium-Proto IX
- PAO:
-
Pheophorbide a oxygenase
- PCBS:
-
Phytochromobilin synthase
- Pchlide:
-
Protochlorophyllide
- pFCC:
-
Primary fluorescent Chl catabolite
- Phein a :
-
Pheophytin a
- Phys:
-
Phytochromes
- PPH:
-
Pheophytinase
- PPO:
-
Protopoiphyrinogen IX oxidase
- Proto IX:
-
Protoporphyrin IX
- RCC:
-
Red Chl catabolite
- RCCR:
-
RCC reductase
- ROS:
-
Reactive oxygen species
- TIC55:
-
Translocon in the inner chloroplast envelope 55
- TPB:
-
Tetrapyrrole biosynthesis
- UROD:
-
Uroporphyrinogen III decarboxylase
- UROS:
-
Uroporphyrinogen III synthetase
- UVR8:
-
UV-B resistance 8
References
Abbas N, Maurya JP, Senapati D et al (2014) Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26:1036–1052. https://doi.org/10.1105/tpc.113.122515
Abbas M, Sharma G, Dambire C et al (2022) An oxygen-sensing mechanism for angiosperm adaptation to altitude. Nature 606:565–569. https://doi.org/10.1038/s41586-022-04740-y
Adhikari ND, Froehlich JE, Strand DD et al (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23:1449–1467. https://doi.org/10.1105/tpc.110.082503
Akmakjian GZ, Riaz N, Guerinot ML (2021) Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3. Proc Natl Acad Sci U S A 118:e2024918118. https://doi.org/10.1073/pnas.2024918118
Allen JF, de Paula WBM, Puthiyaveetil S, Nield J (2011) A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci 16:645–655. https://doi.org/10.1016/j.tplants.2011.10.004
Apel K (1981) The protochlorophyllide holochrome of Barley (Hordeum vulgare L.). Eur J Biochem 120:89–93. https://doi.org/10.1111/j.1432-1033.1981.tb05673.x
Apel K, Hirt H (2004) REACTIVE OXYGEN SPECIES: metabolism, oxidative stress, and signaling transduction. Annu Rev Plant Biol 55:373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Armarego-Marriott T, Sandoval-Ibañez O, Kowalewska Ł (2020) Beyond the darkness: recent lessons from etiolation and de-etiolation studies. J Exp Bot 71:1215–1225. https://doi.org/10.1093/jxb/erz496
Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141:391–396. https://doi.org/10.1104/pp.106.082040
Axelsson E, Lundqvist J, Sawicki A et al (2006) Recessiveness and dominance in barley mutants deficient in Mg-Chelatase Subunit D, an AAA protein involved in chlorophyll biosynthesis. Plant Cell 18:3606–3616. https://doi.org/10.1105/tpc.106.042374
Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59:281–311. https://doi.org/10.1146/annurev.arplant.59.032607.092859
Barry CS, McQuinn RP, Chung M-Y et al (2008) Amino Acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147:179–187. https://doi.org/10.1104/pp.108.118430
Binkert M, Kozma-Bognár L, Terecskei K et al (2014) UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26:4200–4213. https://doi.org/10.1105/tpc.114.130716
Braumann I, Stein N, Hansson M (2014) Reduced chlorophyll biosynthesis in heterozygous barley magnesium chelatase mutants. Plant Physiol Biochem 78:10–14. https://doi.org/10.1016/j.plaphy.2014.02.004
Brzezowski P, Richter AS, Grimm B (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta 1847:968–985. https://doi.org/10.1016/j.bbabio.2015.05.007
Cai R, Zhao Y, Wang Y et al (2014) Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tiss Organ Cult 119:565–577. https://doi.org/10.1007/s11240-014-0556-7
Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70:321–346. https://doi.org/10.1146/annurev-arplant-050718-095919
Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci U S A 108:16475–16480. https://doi.org/10.1073/pnas.1107161108
Cazzonelli CI, Hou X, Alagoz Y et al (2020) A cis-carotene derived apocarotenoid regulates etioplast and chloroplast development. Elife 9:e45310. https://doi.org/10.7554/eLife.45310
Chakraborty N, Tripathy BC (1992) Involvement of singlet oxygen in 5-aminolevulinic acid-induced photodynamic damage of cucumber (Cucumis sativus L.) chloroplasts 1. Plant Physiol 98:7–11. https://doi.org/10.1104/pp.98.1.7
Cheminant S, Wild M, Bouvier F et al (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23:1849–1860. https://doi.org/10.1105/tpc.111.085233
Chen J, Ren G, Kuai B (2016) The mystery of Mendel’s stay-green: magnesium stays chelated in chlorophylls. Mol Plant 9:1556–1558. https://doi.org/10.1016/j.molp.2016.11.004
Chen J, Nolan TM, Ye H et al (2017a) Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in Brassinosteroid-regulated plant growth and drought responses. Plant Cell 29:1425–1439. https://doi.org/10.1105/tpc.17.00364
Chen J, Zhu X, Ren J et al (2017b) Suppressor of overexpression of CO 1 negatively regulates dark-induced leaf degreening and senescence by directly repressing pheophytinase and other senescence-associated genes in Arabidopsis. Plant Physiol 173:1881–1891. https://doi.org/10.1104/pp.16.01457
Cheng M-C, Kathare PK, Paik I, Huq E (2021) Phytochrome signaling networks. Annu Rev Plant Biol 72:217–244. https://doi.org/10.1146/annurev-arplant-080620-024221
Couturier J, Touraine B, Briat J-F et al (2013) The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front Plant Sci 4:259. https://doi.org/10.3389/fpls.2013.00259
Czarnecki O, Grimm B (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot 63:1675–1687. https://doi.org/10.1093/jxb/err437
Czarnecki O, Hedtke B, Melzer M et al (2011) An arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23:4476. https://doi.org/10.1105/tpc.111.086421
de Lucas M, Davière J-M, Rodríguez-Falcón M et al (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451:480–484. https://doi.org/10.1038/nature06520
de Wit M, Galvão VC, Fankhauser C (2016) Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67:513–537. https://doi.org/10.1146/annurev-arplant-043015-112252
Ding Y, Yang S (2022) Surviving and thriving: How plants perceive and respond to temperature stress. Dev Cell 57:947–958. https://doi.org/10.1016/j.devcel.2022.03.010
Dogra V, Kim C (2020) Singlet oxygen metabolism: from genesis to signaling. Front Plant Sci 10:1640. https://doi.org/10.3389/fpls.2019.01640
Ezer D, Shepherd SJK, Brestovitsky A et al (2017) The G-Box transcriptional regulatory code in Arabidopsis. Plant Physiol 175:628–640. https://doi.org/10.1104/pp.17.01086
Farooq M, Wahid A, Basra SMA, Islam-Ud-Din I (2009) Improving water relations and gas exchange with brassinosteroids in rice under drought stress. J Agron Crop Sci 195:262–269. https://doi.org/10.1111/j.1439-037X.2009.00368.x
Feng S, Martinez C, Gusmaroli G et al (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451:475–479. https://doi.org/10.1038/nature06448
Fernández-Milmanda GL, Ballaré CL (2021) Shade avoidance: expanding the color and hormone palette. Trends Plant Sci 26:509–523. https://doi.org/10.1016/j.tplants.2020.12.006
Fusada N, Masuda T, Kuroda H et al (2000) NADPH-protochlorophyllide oxidoreductase in cucumber is encoded by a single gene and its expression is transcriptionally enhanced by illumination. Photosynth Res 64:147–154. https://doi.org/10.1023/A:1006418608647
Gabruk M, Mysliwa-Kurdziel B (2015) Light-dependent protochlorophyllide oxidoreductase: phylogeny, regulation, and catalytic properties. Biochemistry 54:5255–5262. https://doi.org/10.1021/acs.biochem.5b00704
Gao D, Ran C, Zhang Y et al (2022) Effect of different concentrations of foliar iron fertilizer on chlorophyll fluorescence characteristics of iron-deficient rice seedlings under saline sodic conditions. Plant Physiol Biochem 185:112–122. https://doi.org/10.1016/j.plaphy.2022.05.021
Geng D, Chen P, Shen X et al (2018) MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol 178:1296–1309. https://doi.org/10.1104/pp.18.00502
Grajek H, Rydzyński D, Piotrowicz-Cieślak A et al (2020) Cadmium ion-chlorophyll interaction—examination of spectral properties and structure of the cadmium-chlorophyll complex and their relevance to photosynthesis inhibition. Chemosphere 261:127434. https://doi.org/10.1016/j.chemosphere.2020.127434
Guo L, Jiang L, Zhang Y et al (2016) The anaphase-promoting complex initiates zygote division in Arabidopsis through degradation of cyclin B1. Plant J 86:161–174. https://doi.org/10.1111/tpj.13158
Hauenstein M, Christ B, Das A et al (2016) A role for TIC55 as a hydroxylase of phyllobilins, the products of chlorophyll breakdown during plant senescence. Plant Cell 28:2510–2527. https://doi.org/10.1105/tpc.16.00630
Hedtke B, Alawady A, Albacete A et al (2012) Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Mol Biol 78:77–93. https://doi.org/10.1007/s11103-011-9846-1
Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807:977–988. https://doi.org/10.1016/j.bbabio.2010.12.007
Hou Z, Yang Y, Hedtke B, Grimm B (2019) Fluorescence in blue light (FLU) is involved in inactivation and localization of glutamyl-tRNA reductase during light exposure. Plant J 97:517–529. https://doi.org/10.1111/tpj.14138
Ji S, Grimm B, Wang P (2023) Chloroplast SRP43 and SRP54 independently promote thermostability and membrane binding of light-dependent protochlorophyllide oxidoreductases. Plant J 115:1583–1598. https://doi.org/10.1111/tpj.16339
Johnson MP, Goral TK, Duffy CDP et al (2011) Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23:1468–1479. https://doi.org/10.1105/tpc.110.081646
Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91:29–66. https://doi.org/10.1016/S0070-2153(10)91002-8
Khan TA, Fariduddin Q, Yusuf M (2017) Low-temperature stress: is phytohormones application a remedy? Environ Sci Pollut Res Int 24:21574–21590. https://doi.org/10.1007/s11356-017-9948-7
Kindgren P, Norén L, de López J, DB, et al (2012) Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol Plant 5:901–913. https://doi.org/10.1093/mp/ssr112
Kleczkowski LA, Igamberdiev AU (2021) Magnesium signaling in plants. Int J Mol Sci 22:1159. https://doi.org/10.3390/ijms22031159
Kobayashi K, Masuda T (2016) Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front Plant Sci 7:1811. https://doi.org/10.3389/fpls.2016.01811
Kobayashi K, Fujii S, Sasaki D et al (2014) Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front Plant Sci 5:272. https://doi.org/10.3389/fpls.2014.00272
Kovács L, Damkjaer J, Kereïche S et al (2006) Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18:3106–3120. https://doi.org/10.1105/tpc.106.045641
Kruk J, Trebst A (2008) Plastoquinol as a singlet oxygen scavenger in photosystem II. Biochim Biophys Acta 1777:154–162. https://doi.org/10.1016/j.bbabio.2007.10.008
Kruse E, Mock H-P, Grimm B (1995) Coproporphyrinogen III oxidase from barley and tobacco—sequence analysis and initial expression studies. Planta 196:796–803. https://doi.org/10.1007/BF01106776
Kuai B, Chen J, Hörtensteiner S (2018) The biochemistry and molecular biology of chlorophyll breakdown. J Exp Bot 69:751–767. https://doi.org/10.1093/jxb/erx322
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906. https://doi.org/10.1126/science.1079978
Lee B-D, Kim MR, Kang M-Y et al (2017) The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat Commun 8:2259. https://doi.org/10.1038/s41467-017-02476-2
Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16:19–28. https://doi.org/10.1016/j.tplants.2010.08.003
Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60:239–260. https://doi.org/10.1146/annurev.arplant.58.032806.103844
Li Y, Li Q, Beuchat G et al (2021) Combined analyses of translatome and transcriptome in Arabidopsis reveal new players responding to magnesium deficiency. J Integr Plant Biol 63:2075–2092. https://doi.org/10.1111/jipb.13169
Li J-Y, Yang C, Xu J et al (2023) The hot science in rice research: How rice plants cope with heat stress. Plant Cell Environ 46:1087–1103. https://doi.org/10.1111/pce.14509
Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212:563–570. https://doi.org/10.1111/nph.14217
Lin Y-P, Lee T, Tanaka A, Charng Y (2014) Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J 80:14–26. https://doi.org/10.1111/tpj.12611
Liscum E, Nittler P, Koskie K (2020) The continuing arc toward phototropic enlightenment. J Exp Bot 71:1652–1658. https://doi.org/10.1093/jxb/eraa005
Liu Y, Ji X, Nie X et al (2015) Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol 207:692–709. https://doi.org/10.1111/nph.13387
Liu X, Li Y, Zhong S (2017) Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Front Plant Sci 8:1433. https://doi.org/10.3389/fpls.2017.01433
Liu M, Ma W, Su X et al (2022) Mutation in a chlorophyll-binding motif of Brassica ferrochelatase enhances both heme and chlorophyll biosynthesis. Cell Rep 41:111758. https://doi.org/10.1016/j.celrep.2022.111758
Liu W, Chen G, He M et al (2023a) ABI5 promotes heat stress-induced chlorophyll degradation by modulating the stability of MYB44 in cucumber. Hortic Res 10:uhad089. https://doi.org/10.1093/hr/uhad089
Liu XX, Zhu XF, Xue DW et al (2023b) Beyond iron-storage pool: functions of plant apoplastic iron during stress. Trends Plant Sci 28:941–954. https://doi.org/10.1016/j.tplants.2023.03.007
Lu Y (2018) Assembly and transfer of iron-sulfur clusters in the plastid. Front Plant Sci 9:336. https://doi.org/10.3389/fpls.2018.00336
Luquez VM, Guiamét JJ (2001) Effects of the ‘stay green’ genotype GGd1d1d2d2 on leaf gas exchange, dry matter accumulation and seed yield in soybean (Glycine max L. Merr.). Ann Bot 87:313–318. https://doi.org/10.1006/anbo.2000.1324
Ma Y-Y, Shi J-C, Wang D-J et al (2023) A point mutation in the gene encoding Mg-chelatase subunit I influences strawberry leaf color and metabolism. Plant Physiol 192:2737–2755. https://doi.org/10.1093/plphys/kiad247
Maroco JP, Rodrigues ML, Lopes C, Chaves MM (2002) Limitations to leaf photosynthesis in field-grown grapevine under drought - metabolic and modelling approaches. Funct Plant Biol 29:451–459. https://doi.org/10.1071/PP01040
Martín G, Leivar P, Ludevid D et al (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7:11431. https://doi.org/10.1038/ncomms11431
Masuda T, Fusada N, Oosawa N et al (2003) Functional Analysis of Isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol 44:963–974. https://doi.org/10.1093/pcp/pcg128
Meguro M, Ito H, Takabayashi A et al (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23:3442–3453. https://doi.org/10.1105/tpc.111.089714
Nagai S, Koide M, Takahashi S et al (2007) Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol 144:1039–1051. https://doi.org/10.1104/pp.107.100065
Nam H-I, Shahzad Z, Dorone Y et al (2021) Interdependent iron and phosphorus availability controls photosynthesis through retrograde signaling. Nat Commun 12:7211. https://doi.org/10.1038/s41467-021-27548-2
Nath K, Jajoo A, Poudyal RS et al (2013) Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Lett 587:3372–3381. https://doi.org/10.1016/j.febslet.2013.09.015
Nguyen HC, Melo AA, Kruk J et al (2021) Photocatalytic LPOR forms helical lattices that shape membranes for chlorophyll synthesis. Nat Plants 7:437–444. https://doi.org/10.1038/s41477-021-00885-2
Ogura T, Kobayashi NI, Hermans C et al (2020) Short-term magnesium deficiency triggers nutrient retranslocation in Arabidopsis thaliana. Front Plant Sci 11:563. https://doi.org/10.3389/fpls.2020.00563
Paik I, Huq E (2019) Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin Cell Dev Biol 92:114–121. https://doi.org/10.1016/j.semcdb.2019.03.007
Park S-Y, Yu J-W, Park J-S et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19:1649–1664. https://doi.org/10.1105/tpc.106.044891
Pattanayak GK, Tripathy BC (2011) Overexpression of protochlorophyllide oxidoreductase C regulates oxidative stress in Arabidopsis. PLoS ONE 6:e26532. https://doi.org/10.1371/journal.pone.0026532
Peter E, Grimm B (2009) GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol Plant 2:1198–1210. https://doi.org/10.1093/mp/ssp072
Podolec R, Demarsy E, Ulm R (2021) Perception and signaling of ultraviolet-B radiation in plants. Annu Rev Plant Biol 72:793–822. https://doi.org/10.1146/annurev-arplant-050718-095946
Powell ALT, Nguyen CV, Hill T et al (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336:1711–1715. https://doi.org/10.1126/science.1222218
Pružinská A, Tanner G, Anders I et al (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci U S A 100:15259–15264. https://doi.org/10.1073/pnas.2036571100
Qian L, Voss-Fels K, Cui Y et al (2016) Deletion of a stay-green gene associates with adaptive selection in Brassica napus. Mol Plant 9:1559–1569. https://doi.org/10.1016/j.molp.2016.10.017
Rastogi A, Yadav DK, Szymańska R et al (2014) Singlet oxygen scavenging activity of tocopherol and plastochromanol in Arabidopsis thaliana: relevance to photooxidative stress. Plant Cell Environ 37:392–401. https://doi.org/10.1111/pce.12161
Reinbothe C, Bakkouri ME, Buhr F et al (2010) Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci 15:614–624. https://doi.org/10.1016/j.tplants.2010.07.002
Ren G, An K, Liao Y et al (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144:1429–1441. https://doi.org/10.1104/pp.107.100172
Rinalducci S, Pedersen JZ, Zolla L (2004) Formation of radicals from singlet oxygen produced during photoinhibition of isolated light-harvesting proteins of photosystem II. Biochim Biophys Acta 1608:63–73. https://doi.org/10.1016/j.bbabio.2003.10.009
Rodríguez-Celma J, Pan IC, Li W et al (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front Plant Sci 4:276. https://doi.org/10.3389/fpls.2013.00276
Sakuraba Y, Jeong J, Kang M-Y et al (2014a) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5:4636. https://doi.org/10.1038/ncomms5636
Sakuraba Y, Kim D, Kim Y-S et al (2014b) Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett 588:3830–3837. https://doi.org/10.1016/j.febslet.2014.09.018
Santabarbara S, Cazzalini I, Rivadossi A et al (2002) Photoinhibition in vivo and in vitro involves weakly coupled chlorophyll-protein complexes. Photochem Photobiol 75:613–618. https://doi.org/10.1562/0031-8655(2002)0750613PIVAIV2.0.CO2
Santabarbara S, Agostini G, Casazza AP et al (2007) Chlorophyll triplet states associated with Photosystem I and Photosystem II in thylakoids of the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta 1767:88–105. https://doi.org/10.1016/j.bbabio.2006.10.007
Sarkar D, Sharma A, Talukder G (1994) Chlorophyll and chlorophyllin as modifiers of genotoxic effects. Mutat Res 318:239–247. https://doi.org/10.1016/0165-1110(94)90017-5
Sato Y, Morita R, Nishimura M et al (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci U S A 104:14169–14174. https://doi.org/10.1073/pnas.0705521104
Sato Y, Morita R, Katsuma S et al (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57:120–131. https://doi.org/10.1111/j.1365-313X.2008.03670.x
Schelbert S, Aubry S, Burla B et al (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21:767–785. https://doi.org/10.1105/tpc.108.064089
Schmidt SB, Eisenhut M, Schneider A (2020) Chloroplast transition metal regulation for efficient photosynthesis. Trends Plant Sci 25:817–828. https://doi.org/10.1016/j.tplants.2020.03.003
Schwenkert S, Fernie AR, Geigenberger P et al (2022) Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci 27:577–587. https://doi.org/10.1016/j.tplants.2021.12.004
Seaton DD, Smith RW, Song YH et al (2015) Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol Syst Biol 11:776. https://doi.org/10.15252/msb.20145766
Sezgin A, Altuntaş C, Demiralay M et al (2019) Exogenous alpha lipoic acid can stimulate photosystem II activity and the gene expressions of carbon fixation and chlorophyll metabolism enzymes in maize seedlings under drought. J Plant Physiol 232:65–73. https://doi.org/10.1016/j.jplph.2018.11.026
Shan D, Wang C, Song H et al (2021) The MdMEK2-MdMPK6-MdWRKY17 pathway stabilizes chlorophyll levels by directly regulating MdSUFB in apple under drought stress. Plant J 108:814–828. https://doi.org/10.1111/tpj.15480
Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28:2147–2160. https://doi.org/10.1105/tpc.16.00428
Smirnoff N, Arnaud D (2019) Hydrogen peroxide metabolism and functions in plants. New Phytol 221:1197–1214. https://doi.org/10.1111/nph.15488
Song Y, Yang C, Gao S et al (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7:1776–1787. https://doi.org/10.1093/mp/ssu109
Suekawa M, Fujikawa Y, Esaka M (2018) Two G-box–like elements essential to high gene expression of SlAKR4B in tomato leaves. Biosci Biotechnol Biochem 82:425–432. https://doi.org/10.1080/09168451.2018.1429887
Szymańska R, Kruk J (2010) Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in Arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol 51:537–545. https://doi.org/10.1093/pcp/pcq017
Takahashi S, Murata N (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13:178–182. https://doi.org/10.1016/j.tplants.2008.01.005
Tanaka A, Tanaka R (2006) Chlorophyll metabolism. Curr Opin Plant Biol 9:248–255. https://doi.org/10.1016/j.pbi.2006.03.011
Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58:321–346. https://doi.org/10.1146/annurev.arplant.57.032905.105448
Tanaka R, Rothbart M, Oka S et al (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci U S A 107:16721–16725. https://doi.org/10.1073/pnas.1004699107
Tsuchiya Y, Vidaurre D, Toh S et al (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6:741–749. https://doi.org/10.1038/nchembio.435
van Mieghem F, Brettel K, Hillman B et al (1995) Charge recombination reactions in photosystem II. 1. Yields, recombination pathways, and kinetics of the primary pair. Biochemistry 34:4798–4813. https://doi.org/10.1021/bi00014a038
Vandenbussche F, Habricot Y, Condiff AS et al (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49:428–441. https://doi.org/10.1111/j.1365-313X.2006.02973.x
Varshney RK, Sinha P, Singh VK et al (2020) 5Gs for crop genetic improvement. Curr Opin Plant Biol 56:190–196. https://doi.org/10.1016/j.pbi.2019.12.004
Vavilin DV, Vermaas WFJ (2002) Regulation of the tetrapyrrole biosynthetic pathway leading to heme and chlorophyll in plants and cyanobacteria. Physiol Plant 115:9–24. https://doi.org/10.1034/j.1399-3054.2002.1150102.x
Voon CP, Guan X, Sun Y et al (2018) ATP compartmentation in plastids and cytosol of Arabidopsis thaliana revealed by fluorescent protein sensing. Proc Natl Acad Sci USA 115:E10778–E10787. https://doi.org/10.1073/pnas.1711497115
Wang P, Grimm B (2021) Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci 26:484–495. https://doi.org/10.1016/j.tplants.2020.12.005
Wang P, Fouracre J, Kelly S et al (2013) Evolution of GOLDEN2-LIKE gene function in C(3) and C (4) plants. Planta 237:481–495. https://doi.org/10.1007/s00425-012-1754-3
Wang M, Li W, Fang C et al (2018a) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50:1435–1441. https://doi.org/10.1038/s41588-018-0229-2
Wang P, Liang F-C, Wittmann D et al (2018b) Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc Natl Acad Sci USA 115:E3588–E3596. https://doi.org/10.1073/pnas.1719645115
Wang P, Richter AS, Kleeberg JRW et al (2020) Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat Commun 11:1254. https://doi.org/10.1038/s41467-020-14992-9
Wang J, Chen G, Li X et al (2022a) Transcriptome and metabolome analysis of a late-senescent vegetable soybean during seed development provides new insights into degradation of chlorophyll. Antioxidants 11:2480. https://doi.org/10.3390/antiox11122480
Wang T, Liu S, Tian S et al (2022b) Light regulates chlorophyll biosynthesis via ELIP1 during the storage of Chinese cabbage. Sci Rep 12:1–11. https://doi.org/10.1038/s41598-022-15451-9
Wang Z, Zhang Y, Liu Y et al (2023) Calcium-dependent protein kinases CPK21 and CPK23 phosphorylate and activate the iron-regulated transporter IRT1 to regulate iron deficiency in Arabidopsis. Sci China Life Sci 11:2646–2662. https://doi.org/10.1007/s11427-022-2330-4
Waszczak C, Carmody M, Kangasjärvi J (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69:209–236. https://doi.org/10.1146/annurev-arplant-042817-040322
Waters MT, Wang P, Korkaric M et al (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21:1109–1128. https://doi.org/10.1105/tpc.108.065250
Wolf B-C, Isaacson T, Tiwari V et al (2020) Redox regulation of PGRL1 at the onset of low light intensity. Plant J 103:715–725. https://doi.org/10.1111/tpj.14764
Wu S, Li Z, Yang L et al (2016) NON-YELLOWING2 (NYE2), a Close Paralog of NYE1, plays a positive role in Chlorophyll degradation in Arabidopsis. Mol Plant 9:624–627. https://doi.org/10.1016/j.molp.2015.12.016
Xie Z, Wu S, Chen J et al (2019) The C-terminal cysteine-rich motif of NYE1/SGR1 is indispensable for its function in chlorophyll degradation in Arabidopsis. Plant Mol Biol 101:257–268. https://doi.org/10.1007/s11103-019-00902-1
Xu D (2020) COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228:1748–1753. https://doi.org/10.1111/nph.16296
Yamatani H, Ito T, Nishimura K et al (2022) Genetic analysis of chlorophyll synthesis and degradation regulated by BALANCE of CHLOROPHYLL METABOLISM. Plant Physiol 189:419–432. https://doi.org/10.1093/plphys/kiac059
Yu X, Hu S, He C et al (2019) Chlorophyll metabolism in postharvest tea (Camellia sinensis L.) leaves: variations in color values, chlorophyll derivatives, and gene expression levels under different withering treatments. J Agric Food Chem 67:10624–10636. https://doi.org/10.1021/acs.jafc.9b03477
Yuan M, Xu M-Y, Yuan S et al (2010) Light regulation to chlorophyll synthesis and plastid development of the chlorophyll-less golden-leaf privet. J Integr Plant Biol 52:809–816. https://doi.org/10.1111/j.1744-7909.2010.00979.x
Yuan Y, Liu L, Gao Y et al (2022) Comparative analysis of drought-responsive physiological and transcriptome in broomcorn millet (Panicum miliaceum L.) genotypes with contrasting drought tolerance. Ind Crops Prod 177:114498. https://doi.org/10.1016/j.indcrop.2021.114498
Zhang C, Zhang B, Mu B et al (2020) A thylakoid membrane protein functions synergistically with GUN5 in chlorophyll biosynthesis. Plant Commun 1:100094. https://doi.org/10.1016/j.xplc.2020.100094
Zhang W, Willows RD, Deng R et al (2021) Bilin-dependent regulation of chlorophyll biosynthesis by GUN4. Proc Natl Acad Sci USA 118:e2104443118. https://doi.org/10.1073/pnas.2104443118
Zhang H, Zhou J-F, Kan Y et al (2022) A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376:1293–1300. https://doi.org/10.1126/science.abo5721
Zhang T, Zhang R, Zeng X-Y et al (2024) GLK transcription factors accompany ELONGATED HYPOCOTYL5 to orchestrate light-induced seedling development in Arabidopsis. Plant Physiol 194:2400–2421. https://doi.org/10.1093/plphys/kiae002
Zhao J, Zhang X, Guo R et al (2018) Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tiss Organ Cult 132:359–370. https://doi.org/10.1007/s11240-017-1335-z
Zhao Y, Shi H, Pan Y et al (2023) Sensory circuitry controls cytosolic calcium-mediated phytochrome B phototransduction. Cell 186:1230–1243.e14. https://doi.org/10.1016/j.cell.2023.02.011
Zhong S, Zhao M, Shi T et al (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A 106:21431–21436. https://doi.org/10.1073/pnas.0907670106
Zhu J, Cao X, Deng X (2023) Epigenetic and transcription factors synergistically promote the high temperature response in plants. Trends Biochem Sci 48:788–800. https://doi.org/10.1016/j.tibs.2023.06.001
Acknowledgements
Thanks to all the authors for their contributions to this chapter.
Funding
This work was financially supported by The National Natural Science Foundation of China (Grant Numbers: 32102321), the National Key R&D Program of China (No. 2019YFD1000900), the Chongqing Science and Technology Commission (cstc2021jscx-gksbX0010 and cstc2021jcyj-msxmX1156), the Innovation Research Group Funds for Chongqing Universities (CXQT19005), the Chongqing Forestry Administration (YuLinKeYan2022-14), the Characteristic Fruit Industry and Technology System Innovation Team of Chongqing Agriculture and Rural Affairs Commission: No.2020[3]01, and Chongqing Postgraduate Research and Innovation Programme (CYS23213).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.Y., Q.C., and F.X.; writing—original draft preparation, S.Y.; writing—review and editing, Q.C., H.F., F.X., and Q.G.; funding acquisition, Q.G., and G.L. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; the collection, analyses, or interpretation of data; the writing of the manuscript; or the decision to publish the results.
Informed consent statement
Not applicable.
Institutional review board statement
Not applicable.
Additional information
Communicated by Qigao Guo.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yong, S., Chen, Q., Xu, F. et al. Exploring the interplay between angiosperm chlorophyll metabolism and environmental factors. Planta 260, 25 (2024). https://doi.org/10.1007/s00425-024-04437-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04437-8