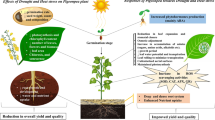
Abstract
Millets stand out as a sustainable crop with the potential to address the issues of food insecurity and malnutrition. These small-seeded, drought-resistant cereals have adapted to survive a broad spectrum of abiotic stresses. Researchers are keen on unravelling the regulatory mechanisms that empower millets to withstand environmental adversities. The aim is to leverage these identified genetic determinants from millets for enhancing the stress tolerance of major cereal crops through genetic engineering or breeding. This review sheds light on transcription factors (TFs) that govern diverse abiotic stress responses and play role in conferring tolerance to various abiotic stresses in millets. Specifically, the molecular functions and expression patterns of investigated TFs from various families, including bHLH, bZIP, DREB, HSF, MYB, NAC, NF-Y and WRKY, are comprehensively discussed. It also explores the potential of TFs in developing stress-tolerant crops, presenting a comprehensive discussion on diverse strategies for their integration.


Similar content being viewed by others
Data availability
The data in the review article are available from the corresponding author upon request.
References
Agarwal PK, Gupta K, Lopato S et al (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot 68:2135–2148
Aida M, IshidaT FH et al (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Ajeesh Krishna TP, Maharajan T, Ceasar SA (2022) Improvement of millets in the post-genomic era. Physiol Mol Biol Plants 28:669–685
Akbudak MA, Filiz E, Kontbay K (2018) DREB2 (dehydration-responsive element-binding protein 2) type transcription factor in sorghum (Sorghum bicolor): genome-wide identification, characterization and expression profiles under cadmium and salt stresses. 3 Biotech 8:1–16
Al-Whaibi MH (2011) Plant heat-shock proteins: a mini review. J King Saud Univ Sci 23:139–150
Ambawat S, Sharma P, Yadav NR et al (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19:307–321
An P, Li X, Liu T et al (2022) The identification of broomcorn millet bZIP transcription factors, which regulate growth and development to enhance stress tolerance and seed germination. Int J Mol Sci 23:6448
Antunes MS, Morey KJ, Tewari-Singh N et al (2009) Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol 5:270
Antunes MS, Morey KJ, Smith JJ et al (2011) Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE 6:e16292
Arakawa H (2016) A method to convert mRNA into a gRNA library for CRISPR/Cas9 editing of any organism. Sci Adv 2:e1600699
Babitha KC, Vemanna RS, Nataraja KN et al (2015) Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 10:e0137098
Bandyopadhyay T, Muthamilarasan M, Prasad M (2017) Millets for next generation climate-smart agriculture. Front Plant Sci 8:1266
Bandyopadhyay T, Swarbreck SM, Jaiswal V (2022) GWAS identifies genetic loci underlying nitrogen responsiveness in the climate resilient C4 model Setaria italica (L.). J Adv Res 42:249–261
Bandyopadhyay T, Singh RK, Ramesh P et al (2023) the promise of millets in the twenty-first century: emphasis on breeding, nutrition, food security and sustainability. J Soil Sci Plant Nutr 23:628–637
Baniwal SK, Bharti K, Chan KY et al (2014) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29:471–487
Baoxiang W, Zhiguang S, Yan L et al (2023) A pervasive phosphorylation cascade modulation of plant transcription factors in response to abiotic stress. Planta 258:73
Bechtold U, Field B (2018) Molecular mechanisms controlling plant growth during abiotic stress. J Exp Bot 69:2753–2758
Bennetzen JL, Schmutz J, Wang H et al (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30:555–561
Bian Z, Gao H, Wang C (2020) NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops. Int J Mol Sci 22:81
Bihani P, Char B, Bhargava S (2011) Transgenic expression of sorghum DREB2 in rice improves tolerance and yield under water limitation. J Agric Sci 149:95–101
Blanc-Mathieu R, Dumas R, Turchi L et al (2023) Plant-TFClass: a structural classification for plant transcription factors. Trends Plant Sci S1360–1385(23):00227–00233
Bouton C, King RC, Chen H et al (2018) Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiol 177:1352–1367
Ceasar A (2022) Genome-editing in millets: current knowledge and future perspectives. Mol Biol Rep 49:773–781
Chanwala J, Satpati S, Dixit A et al (2020) Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom 21:231
Chaudhry S, Sidhu GPS (2022) Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep 41:1–31
Christianson JA, Dennis ES, Llewellyn DJ et al (2010) ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signal Behav 5:428–432
Dou Y, Qin Y, Min D et al (2017) Transcription factor SiNAC18 positively regulates seed germination under drought stress through ABA signaling pathway in foxtail millet (Setaria italic L.). Sci Agric Sin 50:3071–3081
Dudhate A, Shinde H, Yu P et al (2021) Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom 22:1–15
Erpen L, Devi HS, Grosser JW et al (2018) Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult 132:1–25
Fan Y, Lai D, Yang H et al (2021) Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genom 22:778
Fragkostefanakis S, Roeth S, Schleiff E et al (2015) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38:1881–1895
Gawai D, Moharil M, Jadhav P (2017) Differential gene expression in foxtail millet (Setaria italica) under water stress. IJRANSS 5:99–104
Ge L, Dou Y, Li M et al (2019) SiMYB3 in foxtail millet (Setaria italica) confers tolerance to low-nitrogen stress by regulating root growth in transgenic plants. Int J Mol Sci 20:5741
Gonzalez DH (2015) Plant Transcription factors: evolutionary, structural and functional aspects. Elsevier, London, UK
Goold HD, Wright P, Hailstones D (2018) Emerging opportunities for synthetic biology in agriculture. Genes 9:341
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
He M, He CQ, Ding NZ (2018) Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front Plant Sci 9:1771
Hittalmani S, Mahesh HB, Shirke MD et al (2017) Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom 18:16
Hrmova M, Hussain SS (2021) Plant transcription factors involved in drought and associated stresses. Int J Mol Sci 22:5662
Huang D, Sun M, Zhang A et al (2021) Transcriptional changes in pearl millet leaves under heat stress. Genes 12:1716
Inukai S, Kock KH, Bulyk ML (2017) Transcription factor–DNA binding: beyond binding site motifs. Curr Opin Genet Dev 43:110–119
Jiang J, Ma S, Ye N et al (2017) WRKY transcription factors in plant responses to stresses. J Integr Plant Biol 59:86–101
Joshi R, Wani SH, Singh B et al (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029
Kheya SA, Talukder SK, Datta P et al (2023) Millets: the future crops for the tropics-status, challenges and future prospects. Heliyon 9:e22123
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT et al (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet MGG 262:1047–1051
Kosová K, Vítámvás P, Urban MO et al (2015) Biological networks underlying abiotic stress tolerance in temperate crops—a proteomic perspective. Int J Mol Sci 16:20913–20942
Kristensen C, Morant M, Olsen CE et al (2005) Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci 102:1779–1784
Kumar V, Singh B, Kumar Singh R, Sharma N, Muthamilarasan M, Sawant SV, Prasad M (2024) Histone deacetylase 9 interacts with SiHAT3.1 and SiHDA19 to repress dehydration responses through H3K9 deacetylation in foxtail millet. J Exp Bot 75:1098–1111
Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62:4731–4748
Lata C, Bhutty S, Bahadur RP (2011) Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J Exp Bot 62:3387–3401
Li C, Yue J, Wu X et al (2014) An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J Exp Bot 65:5415–5427
Li J, Dong Y, Li C et al (2017) SiASR4, the target gene of SiARDP from Setaria italica, improves abiotic stress adaption in plants. Front Plant Sci 7:2053
Lin CS, Hsu CT, Yang LH et al (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16:1295–1310
Liu Q, Zhang G, Chen S (2001) Structure and regulatory function of plant transcription factors. Chin Sci Bull 46:271–278
Liu N, Xie K, Jia Q et al (2016) Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol 171:1801–1807
Mahesh HB, Shirke MD, Ghodke I et al (2022) Role of inducible promoters and transcription factors in conferring abiotic stress-tolerance in small millets. Omics of Climate resilient small millets. Springer Nature Singapore, Singapore, pp 69–86
Maheshwari P, Kummari D, Palakolanu SR et al (2019) Genome-wide identification and expression profile analysis of nuclear factor Y family genes in Sorghum bicolor L. (Moench). PLoS ONE 14:e0222203
Mantri N, Patade V, Penna S et al (2012) Abiotic stress responses in plants: present and future. Abiotic stress responses in plants. Springer, New York, USA, pp 1–19
Meena RP, Ghosh G, Vishwakarma H et al (2022) Expression of a Pennisetum glaucum gene DREB2A confers enhanced heat, drought and salinity tolerance in transgenic Arabidopsis. Mol Biol Rep 49:7347–7358
Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37:118–125
Mukesh Sankar S, Tara Satyavathi C, Barthakur S et al (2021) Differential modulation of heat-inducible genes across diverse genotypes and molecular cloning of a sHSP from pearl millet [Pennisetum glaucum (L.) R. Br.]. Front Plant Sci 12:659893
Niu J, Guan Y, Yu X et al (2023) SiNF-YC2 regulates early maturity and salt tolerance in Setaria italica. Int J Mol Sci 24:7217
Nover L, Bharti K, Döring P et al (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6:177–189
Nuruzzaman M, Manimekalai R, Sharoni AM et al (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Olsen AN, Ernst HA, Leggio LL et al (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Pandurangaiah M, Gunupuru LR, Veeranagamallaiah G (2016) Molecular cloning and characterization of a novel SiDREB2L gene encoding DRE-binding transcription factor2 protein from Foxtail millet (Setaria italica. L., Cv. Prasad). J Plant Biol Res 5:48–57
Pardey PG, Beddow JM, Hurley TM et al (2014) A bounds analysis of world food futures: global agriculture through to 2050. Aust J Agric Resour Econ 58:571–589
Peng R, Zhang B (2021) Foxtail millet: a new model for C4 plants. Trends Plant Sci 26:199–201
Punia H, Tokas J, Malik A et al (2021) Genome-wide transcriptome profiling, characterization, and functional identification of NAC transcription factors in sorghum under salt stress. Antioxidants 10:1605
Puranik S, Sahu PP, Srivastava PS et al (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Puranik S, Bahadur RP, Srivastava PS et al (2011a) Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Mol Biotechnol 49:138–150
Puranik S, Jha S, Srivastava PS et al (2011b) Comparative transcriptome analysis of contrasting foxtail millet cultivars in response to short-term salinity stress. J Plant Physiol 168:280–287
Puranik S, Sahu PP, Mandal SN et al (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8:e64594
Rahman H, Ramanathan V, Nallathambi J et al (2016) Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol 16:7–20
Ramakrishna C, Singh S, Raghavendrarao S et al (2018) The membrane tethered transcription factor EcbZIP17 from finger millet promotes plant growth and enhances tolerance to abiotic stresses. Sci Rep 8:2148
Ramegowda V, Senthil-Kumar M, Nataraja KN et al (2012) Expression of a finger millet transcription factor, EcNAC1, in tobacco confers abiotic stress-tolerance. PLoS ONE 7:e40397
Rani V, Joshi DC, Joshi P et al (2023) “Millet Models” for harnessing nuclear factor-Y transcription factors to engineer stress tolerance in plants: current knowledge and emerging paradigms. Planta 258:29
Reis SPD, Lima AM, De Souza CRB (2012) Recent molecular advances on downstream plant responses to abiotic stress. Int J Mol Sci 13:8628–8647
Ren T, Wang J, Zhao M et al (2018) Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247:53–68
Rockström J, Williams J, Daily G et al (2017) Sustainable intensification of agriculture for human prosperity and global sustainability. Ambio 46:4–17
Sakuma Y, Liu Q, Dubouzet JG (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sanjari S, Shirzadian-Khorramabad R, Shobbar ZS et al (2019) Systematic analysis of NAC transcription factors’ gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep 38:361–376
Sargent D, Conaty WC, Tissue DT et al (2022) Synthetic biology and opportunities within agricultural crops. J Sustain Agric Environ 1:89–107
Satish L, Rathinapriya P, Muthuramalingam P et al (2020) Overexpression of Erianthus arundinaceus DREB2 transcription factor ameliorates the salinity and drought tolerance in Eleusine coracana cultivars. In: Biology and life sciences forum 4, MDPI.
Shan Z, Jiang Y, Li H et al (2020) Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genom 21:1–13
Shao H, Wang H, Tang X (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6:902
Shinde H, Dudhate A, Tsugama D et al (2019) Pearl millet stress-responsive NAC transcription factor PgNAC21 enhances salinity stress tolerance in Arabidopsis. Plant Physiol Biochem 135:546–553
Shiriga K, Sharma R, Kumar K et al (2014) Genome-wide identification and expression pattern of drought responsive members of the NAC family in maize. Meta Gene 2:407–417
Shiraku ML, Magwanga RO, Zhang Y et al (2022) Late embryogenesis abundant gene LEA3 (Gh_A08G0694) enhances drought and salt stress tolerance in cotton. Int J Biol Macromol 207:700–714
Singh VK, Shukla AK, Singh AK (2019) Impact of climate change on plant-microbe interactions under agroecosystems. Climate change and agricultural ecosystems. Woodland publishing, Cambridge, MA, pp 153–179
Singh RK, Muthamilarasan M, Prasad M (2021a) Biotechnological approaches to dissect climate-resilient traits in millets and their application in crop improvement. J Biotech 327:64–73
Singh S, Chopperla R, Shingote P et al (2021b) Overexpression of EcDREB2A transcription factor from finger millet in tobacco enhances tolerance to heat stress through ROS scavenging. J Biotechnol 336:10–24
Singh S, Koyama H, Bhati KK et al (2021c) The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J Plant Res 134:475–495
Singh RK, Muthamilarasan M, Prasad M (2022a) SiHSFA2e regulated expression of SisHSP21. 9 maintains chloroplast proteome integrity under high temperature stress. Cell Mol Life Sci 79:580
Singh RP, Qidwai S, Singh O et al (2022b) Millets for food and nutritional security in the context of climate resilient agriculture: a review. Int J Plant Soil Sci 31:939–953
Souer E, van Houwelingen A, Kloos D et al (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Su H, Zhang S, Yin Y et al (2015) Genome-wide analysis of NAM-ATAF1, 2-CUC2 transcription factor family in Solanum lycopersicum. J Plant Biochem Biotechnol 24:176–183
Sun M, Huang D, Zhang A et al (2020) Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol 20:1–15
Suzuki N, Rivero RM, Shulaev V et al (2014) Abiotic and biotic stress combinations. New Phytol 203:32–43
Varshney RK, Shi C, Thudi M et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35:969–976
Wang Z, Dane F (2013) NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiol Plant 35:1397–1408
Wang M, Li P, Li C et al (2014) SiLEA14, a novel atypical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biol 14:1–16
Waqas MA, Kaya C, Riaz A et al (2019) Potential mechanisms of abiotic stress tolerance in crop plants induced by thiourea. Front Plant Sci 10:1336
Wu Y, Wen J, Xia Y et al (2022) Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic Res 9:uhac058
Xie LN, Chen M, Min DH et al (2017) The NAC-like transcription factor SiNAC110 in foxtail millet (Setaria italica L.) confers tolerance to drought and high salt stress through an ABA independent signaling pathway. J Integr Agric 16:559–571
Xu W, Tang W, Wang C et al (2020) SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway. Front Plant Sci 11:785
Xu C, Luo M, Sun X et al (2022) SiMYB19 from foxtail millet (Setaria italica) confers transgenic rice tolerance to high salt stress in the field. Int J Mol Sci 23:756
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Genet Genom 238:17–25
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yang Z, Zhang H, Li X et al (2020) A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat Plants 6:1167–1178
Yu Y, Guo DD, Min DH et al (2023) Foxtail millet MYB-like transcription factor SiMYB16 confers salt tolerance in transgenic rice by regulating phenylpropane pathway. Plant Physiol Biochem 195:310–321
Yuan C, Li H, Qin C et al (2020) Foxtail mosaic virus-induced flowering assays in monocot crops. J Exp Bot 71:3012–3023
Yue J, Li C, Liu Y (2014) A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PLoS ONE 9:e100772
Yue H, Wang M, Liu S et al (2016) Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L). BMC Genom 17:343
Zhang G, Liu X, Quan Z et al (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30:549–554
Zhang L, Shu H, Zhang A et al (2017) Foxtail millet WRKY genes and drought stress. The J Agric Sci 155:777–790
Zhang Y, He Z, Qi X et al (2023) Overexpression of MYB-like transcription factor SiMYB30 from foxtail millet (Setaria italica L.) confers tolerance to low nitrogen stress in transgenic rice. Plant Physiol Biochem 196:731–738
Zou C, Li L, Miki D et al (2019) The genome of broomcorn millet. Nat Commun 10:436
Acknowledgements
The authors’ work in this area is supported by J.C. Bose National Fellowship (file no. JBR/2023/000024) as well as research grant from the Ministry of Science and Technology, Gov. of India [grant-CRG/2020/000488 and BT/Ag/Network/Wheat/2019-20]. Panchal, A. acknowledges the Department of Biotechnology (DBT), Government of India, for the research fellowship. The authors are also thankful to DBT-eLibrary Consortium (DeLCON) for providing access to the e-resources. All the figures were made using BioRender (https://biorender.com/).
Author information
Authors and Affiliations
Contributions
Prasad, M., Prusty, A. and Panchal, A. conceptualized and designed the outline of the manuscript. Prusty, A. and Panchal, A. wrote the first draft of the manuscript and prepared the figures. Prusty, A., Panchal, A., Singh, R.K. and Prasad, M. revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest was declared.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Prusty, A., Panchal, A., Singh, R.K. et al. Major transcription factor families at the nexus of regulating abiotic stress response in millets: a comprehensive review. Planta 259, 118 (2024). https://doi.org/10.1007/s00425-024-04394-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04394-2