Abstract
Main conclusion
We identified a typical rice premature senescence leaf mutant 86 (psl86) and exhibited the first global ubiquitination data during rice leaf senescence.
Abstract
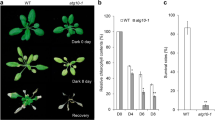
Premature leaf senescence affects the yield and quality of rice, causing irreparable agricultural economic losses. In this study, we reported a rice premature senescence leaf mutant 86 (psl86) in the population lines of rice (Oryza sativa) japonica cultivar ‘Yunyin’ (YY) mutagenized using ethyl methane sulfonate (EMS) treatment. Immunoblotting analysis revealed that a higher ubiquitination level in the psl86 mutant compared with YY. Thus, we performed the proteome and ubiquitylome analyses to identify the differential abundance proteins and ubiquitinated proteins (sites) related to leaf senescence. Among 885 quantified lysine ubiquitination (Kub) sites in 492 proteins, 116 sites in 94 proteins were classified as up-regulated targets and seven sites in six proteins were classified as down-regulated targets at a threshold of 1.5. Proteins with up-regulated Kub sites were mainly enriched in the carbon fixation in photosynthetic organisms, glycolysis/gluconeogenesis and the pentose phosphate pathway. Notably, 14 up-regulated Kub sites in 11 proteins were enriched in the carbon fixation in photosynthetic organism pathway, and seven proteins (rbcL, PGK, GAPA, FBA5, ALDP, CFBP1 and GGAT) were down-regulated, indicating this pathway is tightly regulated by ubiquitination during leaf senescence. To our knowledge, we present the first global data on ubiquitination during rice leaf senescence.








Similar content being viewed by others
Abbreviations
- CBC:
-
Calvin–Benson cycle
- EMS:
-
Ethyl methane sulfonate
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- Kub:
-
Lysine ubiquitination
- psl86 :
-
Premature senescence leaf mutant 86
- YY:
-
Rice japonica cultivar ‘Yunyin
References
Aoyama S, Huarancca RT, Guglielminetti L, Lu Y, Morita Y, Sato T, Yamaguchi J (2014) Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol 55(2):293–305. https://doi.org/10.1093/pcp/pcu002
Asad MA, Zakari SA, Zhao Q, Zhou L, Ye Y, Cheng F (2019) Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: Premature leaf senescence in plants. Int J Mol Sci 20(2):256. https://doi.org/10.3390/ijms20020256
Balazadeh S, Schildhauer J, Araujo WL, Munne-Bosch S, Fernie AR, Proost S, Humbeck K, Mueller-Roeber B (2014) Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: Transcriptomic and metabolomic consequences. J Exp Bot 65(14):3975–3992. https://doi.org/10.1093/jxb/eru119
Chen Y, Xu YY, Luo W, Li WX, Chen N, Zhang DJ, Chong K (2013) The F-Box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol 163(4):1673–1685. https://doi.org/10.1104/pp.113.224527
Chen XL, Xie X, Wu L, Liu C, Zeng L, Zhou X, Luo F, Wang GL, Liu W (2018) Proteomic analysis of ubiquitinated proteins in rice (Oryza sativa) after treatment with pathogen-associated molecular pattern (PAMP) elicitors. Front Plant Sci 9:1064. https://doi.org/10.3389/fpls.2018.01064
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin K, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Rpico A, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Chris Sander C, Schmulevich I, Schwikowsk B, Warner GJ, Ideker T, Bader GD (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Ptotoc 2(10):2366–2382. https://doi.org/10.1038/nprot.2007.324
Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K (2009) Improved visualization of protein consensus sequences by iceLogo. Nat Methods 6(11):786–787. https://doi.org/10.1038/nmeth1109-786
Dametto A, Buffon G, Dos Reis Blasi ÉA, Sperotto RA (2015) Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal Behav 10(9):e1057369. https://doi.org/10.1080/15592324.2015.1057369
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(9): 1–11. http://genomebiology.com/2003/4/9/R60
Gan P, Liu F, Li R, Wang S, Luo J (2019) Chloroplasts- beyond energy capture and carbon fixation: tuning of photosynthesis in response to chilling stress. Int J Mol Sci 20(20):5046. https://doi.org/10.3390/ijms20205046
Gnad F, Gunawardena J, Mann M (2010) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39(Suppl_1):253–260. https://doi.org/10.1093/nar/gkq1159
Guo J, Liu J, Wei Q, Wang R, Yang W, Ma YY, Chen GJ, Yu YX (2017) Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiol 173(1):668–687. https://doi.org/10.1104/pp.16.00795
Han M, Kim CY, Lee J, Lee SK, Jeon JS (2014) OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol Cells 37(7):532–539. https://doi.org/10.14348/molcells.2014.0128
He D, Li M, Damaris RN, Bu C, Xue J, Yang P (2020) Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J 101(6):1430–1447. https://doi.org/10.1111/tpj.14593
Huang J, Yan M, Zhu X, Zhang T, Shen W, Yu P, Wang YT, Sang XC, Yu GL, Zhao BB, He GH (2018) Gene mapping of starch accumulation and premature leaf senescence in the ossac3 mutant of rice. Euphytica 214(10):177. https://doi.org/10.1007/s10681-018-2261-9
Jajic I, Sarna T, Strzalka K (2015) Senescence, stress, and reactive oxygen species. Plants 4(3):393–411. https://doi.org/10.3390/plants4030393
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, F-Quinn A, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Koyama T (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci 5:650. https://doi.org/10.3389/fpls.2014.00650
Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I (2009) Heat shock protein cognate 70–4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21(12):3984–4001. https://doi.org/10.1105/tpc.109.071548
Liang CZ, Wang YQ, Zhu Y, Tang JY, Hu B, Liu LC, Ou SJ, Wu HK, Sun XH, Chu JF, Chu CC (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111(27):10013–10018. https://doi.org/10.1073/pnas.1321568111
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58(1):115–136. https://doi.org/10.1146/annurev.arplant.57.032905.105316
Lim C, Kang K, Shim Y, Sakuraba Y, An G, Paek NC (2020) Rice ethylene response factor 101 promotes leaf senescence through jasmonic acid-mediated regulation of OsNAP and OsMYC2. Front Plant Sci 11:1096. https://doi.org/10.3389/fpls.2020.01096
Lin Y, Tan L, Zhao L, Sun X, Sun C (2016) RLS3, a protein with AAA+ domain localized in chloroplast, sustains leaf longevity in rice. J Integr Plant Biol 58(12):971–982. https://doi.org/10.1111/jipb.12487
Ling Q, Li N, Jarvis P (2017) Chloroplast ubiquitin E3 ligase SP1: does it really function in peroxisomes? Plant Physiol 175(2):586–588. https://doi.org/10.1104/pp.17.00948
Lu JY, Xu YJ, Fan YW, Wang Y, Zhang GF, Liang Y, Jiang CY, Hong B, Gao JP, Ma C (2019) Proteome and ubiquitome changes during rose petal senescence. Int J Mol Sci 20(24):6108. https://doi.org/10.3390/ijms20246108
Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63(2):179–188. https://doi.org/10.1111/j.1365-313X.2010.04233.x
Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300(5617):332–336. https://doi.org/10.1126/science.1080585
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucl Acids Res 35(Suppl_2):182–185. https://doi.org/10.1093/nar/gkm321
Nelson CJ, Millar AH (2015) Protein turnover in plant biology. Nat Plants 1(3):15017. https://doi.org/10.1038/nplants.2015.17
Pak C, Van Doorn WG (2005) Delay of Iris flower senescence by protease inhibitors. New Phytol 165(2):473–480. https://doi.org/10.1111/j.1469-8137.2004.01226.x
Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9(1):51. https://doi.org/10.1186/1472-6807-9-51
Porra R, Thompson W, Kriedemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975(3):384–394
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33(Suppl_2):116–120. https://doi.org/10.1093/nar/gki442
Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59(1):39–51. https://doi.org/10.1111/j.1365-313X.2009.03846.x
Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A (2012) Overproduction of Chl b retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol 53(3):505–517. https://doi.org/10.1093/pcp/pcs006
Saleem MF, Shakoor A, Shahid M, Cheema MA, Shakeel A, Shahid M, Tahir MU, Bilal MF (2018) Removal of early fruit branches as potential regulator of Cry1Ac, antioxidants, senescence and yield in Bt. cotton. Ind Crop Prod 124:885–898. https://doi.org/10.1016/j.indcrop.2018.07.084
Shanmugabalaji V, Chahtane H, Accossato S, Rahire M, Gouzerh G, Lopez-Molina L, Kessler F (2018) Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr Biol 28(16):2616–2623. https://doi.org/10.1016/j.cub.2018.06.006
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Shim KC, Kim SH, Jeon YA, Lee HS, Adeva C, Kang JW, Kim HJ, Tai TH, Ahn SN (2020) A ring-type E3 ubiquitin ligase, OsGW2, controls chlorophyll content and dark-induced senescence in rice. Int J Mol Sci 21(5):1704. https://doi.org/10.3390/ijms21051704
Tamary E, Nevo R, Naveh L, Levin-Zaidman S, Kiss V, Savidor A, Levin Y, Eyal Y, Reich Z, Adam Z (2019) Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct 3(3):e00127. https://doi.org/10.1002/pld3.127
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10(6):385–397. https://doi.org/10.1038/nrm2688
Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteom. https://doi.org/10.1074/mcp.M111.013284
Wang M, Zhang T, Peng H, Luo S, Tan J, Jiang K, Heng YQ, Zhang X, Guo XP, Zheng JK (2018) Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front Plant Sci 9:560. https://doi.org/10.3389/fpls.2018.00560
Woo HR, Kim HJ, Lim PO, Nam HG (2019) Leaf senescence: Systems and dynamics aspects. Annu Rev Plant Biol 70:347–376. https://doi.org/10.1146/annurev-arplant-050718-095859
Xie X, Kang H, Liu W, Wang GL (2015) Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J Proteome Res 14(5):2017–2025. https://doi.org/10.1021/pr5009724
Xu FQ, Xue HW (2019) The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ 42(10):2931–2944. https://doi.org/10.1111/pce.13633
Xu X, Guo K, Liang W, Chen Q, Shi J, Shen B (2018) Quantitative proteomics analysis of proteins involved in leaf senescence of rice (Oryza sativa L). Plant Growth Regul 84(2):341–349. https://doi.org/10.1007/s10725-017-0345-5
Zhang N, Xu J, Liu X, Liang WX, Xin MM, Du JK, Hu ZR, Peng HR, Guo WR, Ni ZF, Sun QX, Yao YY (2019) Identification of HSP90C as a substrate of E3 ligase TaSAP5 through ubiquitylome profiling. Plant Sci 287:110170. https://doi.org/10.1016/j.plantsci.2019.110170
Zhou D, Li T, Yang Y, Qu Z, Ouyang L, Jiang Z, Lin X, Zhu C, Peng L, Fu J, Peng X, Bian J, Tang W, Xu J, He H (2020) OsPLS4 is involved in cuticular wax biosynthesis and affects leaf senescence in rice. Front Plant Sci 11:782. https://doi.org/10.3389/fpls.2020.00782
Acknowledgements
This work was supported by the National Key R&D Program Foundation of China (Grant no. 2016YFD0300508), Key program of Science and Technology in Fujian province, China (No. 2020NZ08016), and The Special Foundation of Non-Profit Research Institutes of Fujian Province (Grant no. 2018R1021-5).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflicts of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2021_3793_MOESM7_ESM.xlsx
Table S7 Kyoto Encyclopedia of Genes and Genomes enrichment analysis of differentially modified sites (proteins) (XLSX 65 kb)
425_2021_3793_MOESM9_ESM.xlsx
Table S9 Differentially expressed proteins and modified sites (proteins) in the carbon fixation in photosynthetic organisms pathway (map00710) (XLSX 24 kb)
Rights and permissions
About this article
Cite this article
Yu, X., Zhu, Y., Xie, Y. et al. Ubiquitylomes and proteomes analyses provide a new interpretation of the molecular mechanisms of rice leaf senescence. Planta 255, 43 (2022). https://doi.org/10.1007/s00425-021-03793-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03793-z