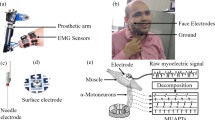
Abstract
Muscle synergy analysis is increasingly used in domains such as neurosciences, robotics, rehabilitation or sport sciences to analyze and better understand motor coordination. The analysis uses dimensionality reduction techniques to identify regularities in spatial, temporal or spatio-temporal patterns of multiple muscle activation. Recent studies have pointed out variability in outcomes associated with the different methodological options available and there was a need to clarify several aspects of the analysis methodology. While synergy analysis appears to be a robust technique, it remain a statistical tool and is, therefore, sensitive to the amount and quality of input data (EMGs). In particular, attention should be paid to EMG amplitude normalization, baseline noise removal or EMG filtering which may diminish or increase the signal-to-noise ratio of the EMG signal and could have major effects on synergy estimates. In order to robustly identify synergies, experiments should be performed so that the groups of muscles that would potentially form a synergy are activated with a sufficient level of activity, ensuring that the synergy subspace is fully explored. The concurrent use of various synergy formulations-spatial, temporal and spatio-temporal synergies- should be encouraged. The number of synergies represents either the dimension of the spatial structure or the number of independent temporal patterns, and we observed that these two aspects are often mixed in the analysis. To select a number, criteria based on noise estimates, reliability of analysis results, or functional outcomes of the synergies provide interesting substitutes to criteria solely based on variance thresholds.






Similar content being viewed by others
Abbreviations
- EMG:
-
Electromyography
- ICA:
-
Independent component analysis
- iEMG:
-
Integrated EMG
- NNMF:
-
Non-negative matrix factorization
- PCA:
-
Principal component analysis
- pNMF:
-
Projective non-negative matrix factorization
- RMS:
-
Root mean square
- SNR:
-
Signal to noise ratio
- SSE:
-
Sum of squared errors
- SST:
-
Total sum of square
- SVD:
-
Singular value decomposition
- VAF:
-
Variance accounted for
References
Ajiboye AB, Weir RF (2009) Muscle synergies as a predictive framework for the EMG patterns of new hand postures. J Neural Eng 6(3):036004. https://doi.org/10.1088/1741-2560/6/3/036004
Allen JL, McKay JL, Sawers A, Hackney ME, Ting LH (2017) Increased neuromuscular consistency in gait and balance after partnered, dance-based rehabilitation in Parkinson’s disease. J Neurophysiol 118(1):363–373. https://doi.org/10.1152/jn.00813.2016
Banks CL, Pai MM, McGuirk TE, Fregly BJ, Patten C (2017) Methodological choices in muscle synergy analysis impact differentiation of physiological characteristics following stroke. Front Comput Neurosci 11:78
Barradas VR, Kutch JJ, Kawase T, Koike Y, Schweighofer N (2020) When 90% of the variance is not enough: residual EMG from muscle synergy extraction influences task performance. J Neurophysiol. https://doi.org/10.1152/jn.00472.2019
Barroso FO, Torricelli D, Moreno JC, Taylor J, Gomez-Soriano J, Bravo-Esteban E, Piazza S, Santos C, Pons JL (2014) Shared muscle synergies in human walking and cycling. J Neurophysiol 112(8):1984–1998. https://doi.org/10.1152/jn.00220.2014
Bell AJ, Sejnowski TJ (1995) Blind separation and blind deconvolution: an information-theoretic approach. In: 1995 International conference on acoustics, speech, and signal processing, 1995. IEEE, pp 3415–3418
Berger DJ, Masciullo M, Molinari M, Lacquaniti F, d’Avella A (2020) Does the cerebellum shape the spatiotemporal organization of muscle patterns? J Neurophysiol, Insights from subjects with cerebellar ataxias. https://doi.org/10.1152/jn.00657.2018
Besomi M, Hodges PW, Clancy EA, Van Dieën J, Hug F, Lowery M, Merletti R, Søgaard K, Wrigley T, Besier T, Carson RG, Disselhorst-Klug C, Enoka RM, Falla D, Farina D, Gandevia S, Holobar A, Kiernan MC, McGill K, Perreault E, Rothwell JC, Tucker K (2020) Consensus for experimental design in electromyography (CEDE) project: amplitude normalization matrix. J Electromyogr Kinesiol 53:102438. https://doi.org/10.1016/j.jelekin.2020.102438
Bizzi E, Cheung VC (2013) The neural origin of muscle synergies. Front Comput Neurosci 7:51
Boonstra TW, Danna-Dos-Santos A, Xie HB, Roerdink M, Stins JF, Breakspear M (2015) Muscle networks: connectivity analysis of EMG activity during postural control. Sci Rep 5:17830. https://doi.org/10.1038/srep17830
Boutsidis C, Gallopoulos E (2008) SVD based initialization: a head start for nonnegative matrix factorization. Pattern Recogn 41(4):1350–1362
Bruton M, O’Dwyer N (2018) Synergies in coordination: a comprehensive overview of neural, computational, and behavioral approaches. J Neurophysiol 120(6):2761–2774. https://doi.org/10.1152/jn.00052.2018
Burkholder TJ, van Antwerp KW (2013) Practical limits on muscle synergy identification by non-negative matrix factorization in systems with mechanical constraints. Med Biol Eng Comput 51(1–2):187–196. https://doi.org/10.1007/s11517-012-0983-8
Campanini I, Merlo A, Degola P, Merletti R, Vezzosi G, Farina D (2007) Effect of electrode location on EMG signal envelope in leg muscles during gait. J Electromyogr Kinesiol 17(4):515–526. https://doi.org/10.1016/j.jelekin.2006.06.001
Cappellini G, Ivanenko YP, Martino G, MacLellan MJ, Sacco A, Morelli D, Lacquaniti F (2016) Immature spinal locomotor output in children with cerebral palsy. Front Physiol 7:478. https://doi.org/10.3389/fphys.2016.00478
Castellini C, van der Smagt P (2013) Evidence of muscle synergies during human grasping. Biol Cybern 107(2):233–245. https://doi.org/10.1007/s00422-013-0548-4
Charissou C, Amarantini D, Baurès R, Berton E, Vigouroux L (2017) Effects of hand configuration on muscle force coordination, co-contraction and concomitant intermuscular coupling during maximal isometric flexion of the fingers. Eur J Appl Physiol 117(11):2309–2320
Cheung VC, d’Avella A, Tresch MC, Bizzi E (2005) Central and sensory contributions to the activation and organization of muscle synergies during natural motor behaviors. J Neurosci 25(27):6419–6434. https://doi.org/10.1523/jneurosci.4904-04.2005
Cheung VC, Piron L, Agostini M, Silvoni S, Turolla A, Bizzi E (2009) Stability of muscle synergies for voluntary actions after cortical stroke in humans. Proc Natl Acad Sci USA 106(46):19563–19568. https://doi.org/10.1073/pnas.0910114106
Cheung VC, Turolla A, Agostini M, Silvoni S, Bennis C, Kasi P, Paganoni S, Bonato P, Bizzi E (2012) Muscle synergy patterns as physiological markers of motor cortical damage. Proc Natl Acad Sci USA 109(36):14652–14656. https://doi.org/10.1073/pnas.1212056109
Chia Bejarano N, Pedrocchi A, Nardone A, Schieppati M, Baccinelli W, Monticone M, Ferrigno G, Ferrante S (2017) Tuning of muscle synergies during walking along rectilinear and curvilinear trajectories in humans. Ann Biomed Eng 45(5):1204–1218. https://doi.org/10.1007/s10439-017-1802-z
Chiovetto E, Endres D, d’Avella A, Giese M (2014) Model selection for the extraction of EMG synergies. dim 2:1
Chiovetto E, d'Avella A, Giese M (2016) A unifying framework for the identification of motor primitives. arXiv preprint arXiv:160306879
Choi S, Cichocki A, Park H-M, Lee S-Y (2005) Blind source separation and independent component analysis: a review. Neural Inf Process Lett Rev 6(1):1–57
Clark DJ, Ting LH, Zajac FE, Neptune RR, Kautz SA (2010) Merging of healthy motor modules predicts reduced locomotor performance and muscle coordination complexity post-stroke. J Neurophysiol 103(2):844–857
d’Avella A, Tresch M (2006) Muscle synergies for motor control. In Handbook of neural engineering. Wiley, New York, pp 449–465
d’Avella A, Saltiel P, Bizzi E (2003) Combinations of muscle synergies in the construction of a natural motor behavior. Nat Neurosci 6(3):300–308. https://doi.org/10.1038/nn1010
d’Avella A, Portone A, Fernandez L, Lacquaniti F (2006) Control of fast-reaching movements by muscle synergy combinations. J Neurosci 26(30):7791–7810. https://doi.org/10.1523/jneurosci.0830-06.2006
Davis BL, Vaughan CL (1993) Phasic behavior of EMG signals during gait: use of multivariate statistics. J Electromyogr Kinesiol 3(1):51–60. https://doi.org/10.1016/1050-6411(93)90023-p
De Luca CJ, Gilmore LD, Kuznetsov M, Roy SH (2010) Filtering the surface EMG signal: movement artifact and baseline noise contamination. J Biomech 43(8):1573–1579. https://doi.org/10.1016/j.jbiomech.2010.01.027
De Marchis C, Severini G, Castronovo AM, Schmid M, Conforto S (2015) Intermuscular coherence contributions in synergistic muscles during pedaling. Exp Brain Res 233(6):1907–1919. https://doi.org/10.1007/s00221-015-4262-4
Del Vecchio A, Germer CM, Elias LA, Fu Q, Fine J, Santello M, Farina D (2019) The human central nervous system transmits common synaptic inputs to distinct motor neuron pools during non-synergistic digit actions. J Physiol 597(24):5935–5948. https://doi.org/10.1113/jp278623
Delis I, Berret B, Pozzo T, Panzeri S (2013) A methodology for assessing the effect of correlations among muscle synergy activations on task-discriminating information. Front Comput Neurosci 7:54. https://doi.org/10.3389/fncom.2013.00054
Delis I, Panzeri S, Pozzo T, Berret B (2014) A unifying model of concurrent spatial and temporal modularity in muscle activity. J Neurophysiol 111(3):675–693. https://doi.org/10.1152/jn.00245.2013
Delis I, Hilt P, Pozzo T, Panzeri S, Berret B (2018) Characterization of whole-body muscle activity during reaching movements using space-by-time modularity and functional similarity analysis. In: Proceedings of the 10th hellenic conference on artificial intelligence, 2018. pp 1–7
Drew T, Kalaska J, Krouchev N (2008) Muscle synergies during locomotion in the cat: a model for motor cortex control. J Physiol 586(5):1239–1245. https://doi.org/10.1113/jphysiol.2007.146605
Ebied A, Kinney-Lang E, Spyrou L, Escudero J (2018) Evaluation of matrix factorisation approaches for muscle synergy extraction. Med Eng Phys 57:51–60. https://doi.org/10.1016/j.medengphy.2018.04.003
Esmaeili J, Maleki A (2020) Muscle coordination analysis by time-varying muscle synergy extraction during cycling across various mechanical conditions. Biocybern Biomed Eng 40(1):90–99
Farina D, Merletti R, Enoka RM (2014) The extraction of neural strategies from the surface EMG: an update. J Appl Physiol (1985) 117(11):1215–1230. https://doi.org/10.1152/japplphysiol.00162.2014
Farina D, Negro F, Muceli S, Enoka RM (2016) Principles of motor unit physiology evolve with advances in technology. Physiology (Bethesda) 31(2):83–94. https://doi.org/10.1152/physiol.00040.2015
Flash T, Hochner B (2005) Motor primitives in vertebrates and invertebrates. Curr Opin Neurobiol 15(6):660–666
Frere J (2017) Spectral properties of multiple myoelectric signals: new insights into the neural origin of muscle synergies. Neuroscience 355:22–35. https://doi.org/10.1016/j.neuroscience.2017.04.039
Fuglevand AJ, Winter DA, Patla AE, Stashuk D (1992) Detection of motor unit action potentials with surface electrodes: influence of electrode size and spacing. Biol Cybern 67(2):143–153. https://doi.org/10.1007/bf00201021
Gallego JA, Perich MG, Miller LE, Solla SA (2017) Neural manifolds for the control of movement. Neuron 94(5):978–984
Ghislieri M, Agostini V, Knaflitz M (2020) Muscle synergies extracted using principal activations: improvement of robustness and interpretability. IEEE Trans Neural Syst Rehabil Eng 28(2):453–460. https://doi.org/10.1109/tnsre.2020.2965179
Giszter SF (2015) Motor primitives—new data and future questions. Curr Opin Neurobiol 33:156–165
Giszter S, Patil V, Hart C (2007) Primitives, premotor drives, and pattern generation: a combined computational and neuroethological perspective. Prog Brain Res 165:323–346
Gizzi L, Nielsen JF, Felici F, Ivanenko YP, Farina D (2011) Impulses of activation but not motor modules are preserved in the locomotion of subacute stroke patients. J Neurophysiol 106(1):202–210. https://doi.org/10.1152/jn.00727.2010
Heales LJ, Hug F, MacDonald DA, Vicenzino B, Hodges PW (2016) Is synergistic organisation of muscle coordination altered in people with lateral epicondylalgia? A case–control study. Clin Biomech (Bristol, Avon) 35:124–131. https://doi.org/10.1016/j.clinbiomech.2016.04.017
Hinnekens E, Berret B, Do MC, Teulier C (2020) Modularity underlying the performance of unusual locomotor tasks inspired by developmental milestones. J Neurophysiol 123(2):496–510. https://doi.org/10.1152/jn.00662.2019
Hirashima M, Oya T (2016) How does the brain solve muscle redundancy? Filling the gap between optimization and muscle synergy hypotheses. Neurosci Res 104:80–87
Hogan N, Sternad D (2013) Dynamic primitives in the control of locomotion. Front Comput Neurosci 7:71
Holdefer R, Miller L (2002) Primary motor cortical neurons encode functional muscle synergies. Exp Brain Res 146(2):233–243
Hug F, Turpin NA, Couturier A, Dorel S (2011) Consistency of muscle synergies during pedaling across different mechanical constraints. J Neurophysiol 106(1):91–103. https://doi.org/10.1152/jn.01096.2010
Hug F, Turpin NA, Dorel S, Guevel A (2012) Smoothing of electromyographic signals can influence the number of extracted muscle synergies. Clin Neurophysiol 123(9):1895–1896. https://doi.org/10.1016/j.clinph.2012.01.015
Hug F, Vogel C, Tucker K, Dorel S, Deschamps T, Le Carpentier E, Lacourpaille L (2019) Individuals have unique muscle activation signatures as revealed during gait and pedaling. J Appl Physiol (1985) 127(4):1165–1174. https://doi.org/10.1152/japplphysiol.01101.2018
Israely S, Leisman G, Machluf C, Shnitzer T, Carmeli E (2017) Direction modulation of muscle synergies in a hand-reaching task. IEEE Trans Neural Syst Rehabil Eng 25(12):2427–2440. https://doi.org/10.1109/tnsre.2017.2769659
Ivanenko YP, Poppele RE, Lacquaniti F (2004) Five basic muscle activation patterns account for muscle activity during human locomotion. J Physiol 556(Pt 1):267–282. https://doi.org/10.1113/jphysiol.2003.057174
Ivanenko YP, Cappellini G, Dominici N, Poppele RE, Lacquaniti F (2005) Coordination of locomotion with voluntary movements in humans. J Neurosci 25(31):7238–7253. https://doi.org/10.1523/jneurosci.1327-05.2005
Jackson DA (1993) Stopping rules in principal components analysis: a comparison of heuristical and statistical approaches. Ecology 74(8):2204–2214
Kargo WJ, Giszter SF (2008) Individual premotor drive pulses, not time-varying synergies, are the units of adjustment for limb trajectories constructed in spinal cord. J Neurosci 28(10):2409–2425. https://doi.org/10.1523/jneurosci.3229-07.2008
Kargo WJ, Nitz DA (2003) Early skill learning is expressed through selection and tuning of cortically represented muscle synergies. J Neurosci 23(35):11255–11269
Kerkman JN, Daffertshofer A, Gollo LL, Breakspear M, Boonstra TW (2018) Network structure of the human musculoskeletal system shapes neural interactions on multiple time scales. Sci Adv. https://doi.org/10.1126/sciadv.aat0497
Kieliba P, Tropea P, Pirondini E, Coscia M, Micera S, Artoni F (2018) How are muscle synergies affected by electromyography pre-processing? IEEE Trans Neural Syst Rehabil Eng 26(4):882–893. https://doi.org/10.1109/tnsre.2018.2810859
Kim Y, Bulea TC, Damiano DL (2016) Novel methods to enhance precision and reliability in muscle synergy identification during walking. Front Hum Neurosci 10:455. https://doi.org/10.3389/fnhum.2016.00455
Kim M, Kim Y, Kim H, Yoon B (2018a) Specific muscle synergies in national elite female ice hockey players in response to unexpected external perturbation. J Sports Sci 36(3):319–325. https://doi.org/10.1080/02640414.2017.1306090
Kim Y, Bulea TC, Damiano DL (2018b) Children with cerebral palsy have greater stride-to-stride variability of muscle synergies during gait than typically developing children: implications for motor control complexity. Neurorehabil Neural Repair 32(9):834–844. https://doi.org/10.1177/1545968318796333
Klein CS, Brooks D, Richardson D, McIlroy WE, Bayley MT (2010) Voluntary activation failure contributes more to plantar flexor weakness than antagonist coactivation and muscle atrophy in chronic stroke survivors. J Appl Physiol (1985) 109(5):1337–1346. https://doi.org/10.1152/japplphysiol.00804.2009
Klein Breteler MD, Simura KJ, Flanders M (2007) Timing of muscle activation in a hand movement sequence. Cereb Cortex 17(4):803–815. https://doi.org/10.1093/cercor/bhk033
Krishnamoorthy V, Latash ML, Scholz JP, Zatsiorsky VM (2004) Muscle modes during shifts of the center of pressure by standing persons: effect of instability and additional support. Exp Brain Res 157(1):18–31. https://doi.org/10.1007/s00221-003-1812-y
Krouchev N, Kalaska JF, Drew T (2006) Sequential activation of muscle synergies during locomotion in the intact cat as revealed by cluster analysis and direct decomposition. J Neurophysiol 96(4):1991–2010. https://doi.org/10.1152/jn.00241.2006
Lacquaniti F, Ivanenko YP, Zago M (2012) Patterned control of human locomotion. J Physiol 590(10):2189–2199
Laine CM, Valero-Cuevas FJ (2017) Intermuscular coherence reflects functional coordination. J Neurophysiol 118(3):1775–1783. https://doi.org/10.1152/jn.00204.2017
Lambert-Shirzad N, Van der Loos HM (2017) On identifying kinematic and muscle synergies: a comparison of matrix factorization methods using experimental data from the healthy population. J Neurophysiol 117(1):290–302
Latash ML (2020) On primitives in motor control. Mot Control 24(2):318–346
Law LF, Krishnan C, Avin K (2011) Modeling nonlinear errors in surface electromyography due to baseline noise: a new methodology. J Biomech 44(1):202–205. https://doi.org/10.1016/j.jbiomech.2010.09.008
Lee WA (1984) Neuromotor synergies as a basis for coordinated intentional action. J Mot Behav 16(2):135–170
Lee DD, Seung HS (1999) Learning the parts of objects by non-negative matrix factorization. Nature 401(6755):788–791. https://doi.org/10.1038/44565
Lee DD, Seung HS (2001) Algorithms for non-negative matrix factorization. In Advances in neural information processing systems. Springer, New York, pp 556–562
Levine AJ, Hinckley CA, Hilde KL, Driscoll SP, Poon TH, Montgomery JM, Pfaff SL (2014) Identification of a cellular node for motor control pathways. Nat Neurosci 17(4):586–593. https://doi.org/10.1038/nn.3675
Lindstroem R, Graven-Nielsen T, Falla D (2012) Current pain and fear of pain contribute to reduced maximum voluntary contraction of neck muscles in patients with chronic neck pain. Arch Phys Med Rehabil 93(11):2042–2048. https://doi.org/10.1016/j.apmr.2012.04.014
Lunardini F, Casellato C, Bertucco M, Sanger TD, Pedrocchi A (2017) Children with and without dystonia share common muscle synergies while performing writing tasks. Ann Biomed Eng 45(8):1949–1962. https://doi.org/10.1007/s10439-017-1838-0
Maclellan MJ, Ivanenko YP, Massaad F, Bruijn SM, Duysens J, Lacquaniti F (2014) Muscle activation patterns are bilaterally linked during split-belt treadmill walking in humans. J Neurophysiol 111(8):1541–1552. https://doi.org/10.1152/jn.00437.2013
Manickaraj N, Bisset LM, Devanaboyina V, Kavanagh JJ (2017) Chronic pain alters spatiotemporal activation patterns of forearm muscle synergies during the development of grip force. J Neurophysiol 118(4):2132–2141. https://doi.org/10.1152/jn.00210.2017
Martinez-Valdes E, Negro F, Falla D, De Nunzio AM, Farina D (2018) Surface electromyographic amplitude does not identify differences in neural drive to synergistic muscles. J Appl Physiol (1985) 124(4):1071–1079. https://doi.org/10.1152/japplphysiol.01115.2017
Milner-Brown HS, Stein RB (1975) The relation between the surface electromyogram and muscular force. J Physiol 246(3):549–569. https://doi.org/10.1113/jphysiol.1975.sp010904
Muceli S, Boye AT, d’Avella A, Farina D (2010) Identifying representative synergy matrices for describing muscular activation patterns during multidirectional reaching in the horizontal plane. J Neurophysiol 103(3):1532–1542. https://doi.org/10.1152/jn.00559.2009
Muceli S, Falla D, Farina D (2014) Reorganization of muscle synergies during multidirectional reaching in the horizontal plane with experimental muscle pain. J Neurophysiol 111(8):1615–1630. https://doi.org/10.1152/jn.00147.2013
Mussa-Ivaldi FA, Solla SA (2004) Neural primitives for motion control. IEEE J Ocean Eng 29(3):640–650
Oliveira AS, Gizzi L, Farina D, Kersting UG (2014) Motor modules of human locomotion: influence of EMG averaging, concatenation, and number of step cycles. Front Hum Neurosci 8:335. https://doi.org/10.3389/fnhum.2014.00335
Overduin SA, d'Avella A, Roh J, Carmena JM, Bizzi E (2015) Representation of muscle synergies in the primate brain. J Neurosci 35(37):12615–12624. https://doi.org/10.1523/jneurosci.4302-14.2015
Rabbi MF, Pizzolato C, Lloyd DG, Carty CP, Devaprakash D, Diamond LE (2020) Non-negative matrix factorisation is the most appropriate method for extraction of muscle synergies in walking and running. Sci Rep 10(1):8266. https://doi.org/10.1038/s41598-020-65257-w
Ranganathan R, Krishnan C (2012) Extracting synergies in gait: using EMG variability to evaluate control strategies. J Neurophysiol 108(5):1537–1544
Rickard S, Cichocki A (2008) When is non-negative matrix decomposition unique? In: 2008 42nd Annual conference on information sciences and systems, 2008. IEEE, pp 1091–1092
Roh J, Rymer WZ, Beer RF (2012) Robustness of muscle synergies underlying three-dimensional force generation at the hand in healthy humans. J Neurophysiol 107(8):2123–2142. https://doi.org/10.1152/jn.00173.2011
Roh J, Rymer WZ, Perreault EJ, Yoo SB, Beer RF (2013) Alterations in upper limb muscle synergy structure in chronic stroke survivors. J Neurophysiol 109(3):768–781. https://doi.org/10.1152/jn.00670.2012
Saito A, Tomita A, Ando R, Watanabe K, Akima H (2018) Similarity of muscle synergies extracted from the lower limb including the deep muscles between level and uphill treadmill walking. Gait Posture 59:134–139. https://doi.org/10.1016/j.gaitpost.2017.10.007
Santuz A, Ekizos A, Janshen L, Baltzopoulos V, Arampatzis A (2017) On the methodological implications of extracting muscle synergies from human locomotion. Int J Neural Syst 27(5):1750007. https://doi.org/10.1142/s0129065717500071
Sartori M, Gizzi L, Lloyd DG, Farina D (2013) A musculoskeletal model of human locomotion driven by a low dimensional set of impulsive excitation primitives. Front Comput Neurosci 7:79. https://doi.org/10.3389/fncom.2013.00079
Sawers A, Allen JL, Ting LH (2015) Long-term training modifies the modular structure and organization of walking balance control. J Neurophysiol 114(6):3359–3373. https://doi.org/10.1152/jn.00758.2015
Scano A, Dardari L, Molteni F, Giberti H, Tosatti LM, d’Avella A (2019) A comprehensive spatial mapping of muscle synergies in highly variable upper-limb movements of healthy subjects. Front Physiol 10:1231. https://doi.org/10.3389/fphys.2019.01231
Severini G, De Marchis C (2018) Effect of SNR normalization on the estimation of muscle synergies from EMG datasets. In: 2018 IEEE international symposium on medical measurements and applications (MeMeA), 2018. IEEE, pp 1–5
Shiavi R, Frigo C, Pedotti A (1998) Electromyographic signals during gait: criteria for envelope filtering and number of strides. Med Biol Eng Comput 36(2):171–178. https://doi.org/10.1007/bf02510739
Shmuelof L, Krakauer JW, Mazzoni P (2012) How is a motor skill learned? Change and invariance at the levels of task success and trajectory control. J Neurophysiol 108(2):578–594. https://doi.org/10.1152/jn.00856.2011
Shuman BR, Schwartz MH, Steele KM (2017) Electromyography data processing impacts muscle synergies during gait for unimpaired children and children with cerebral palsy. Front Comput Neurosci 11:50. https://doi.org/10.3389/fncom.2017.00050
Shuman BR, Goudriaan M, Desloovere K, Schwartz MH, Steele KM (2019) Muscle synergies demonstrate only minimal changes after treatment in cerebral palsy. J Neuroeng Rehabil 16(1):46. https://doi.org/10.1186/s12984-019-0502-3
Singh RE, Iqbal K, White G, Hutchinson TE (2018) A systematic review on muscle synergies: from building blocks of motor behavior to a neurorehabilitation tool. Appl Bionics Biomech. https://doi.org/10.1155/2018/3615368
Soomro MH, Conforto S, Giunta G, Ranaldi S, De Marchis C (2018) Comparison of initialization techniques for the accurate extraction of muscle synergies from myoelectric signals via nonnegative matrix factorization. Appl Bionics Biomech
Sousa AS, Tavares JMR (2012) Surface electromyographic amplitude normalization methods: a review. Electromyogr New Dev Proced Appl
Steele KM, Tresch MC, Perreault EJ (2013) The number and choice of muscles impact the results of muscle synergy analyses. Front Comput Neurosci 7:105. https://doi.org/10.3389/fncom.2013.00105
Steele KM, Rozumalski A, Schwartz MH (2015a) Muscle synergies and complexity of neuromuscular control during gait in cerebral palsy. Dev Med Child Neurol 57(12):1176–1182
Steele KM, Tresch MC, Perreault EJ (2015b) Consequences of biomechanically constrained tasks in the design and interpretation of synergy analyses. J Neurophysiol 113(7):2102–2113
Takei T, Confais J, Tomatsu S, Oya T, Seki K (2017) Neural basis for hand muscle synergies in the primate spinal cord. Proc Natl Acad Sci USA 114(32):8643–8648. https://doi.org/10.1073/pnas.1704328114
Ting LH, Chvatal SA (2010) Decomposing muscle activity in motor tasks: methods and interpretation. In: Danion F, Latash M (eds) Motor control:theories, experiments, and applications. Oxford University Press, Oxford, pp 102–138
Ting LH, Macpherson JM (2005) A limited set of muscle synergies for force control during a postural task. J Neurophysiol 93(1):609–613. https://doi.org/10.1152/jn.00681.2004
Togo S, Imamizu H (2017) Empirical evaluation of voluntarily activatable muscle synergies. Front Comput Neurosci 11:82. https://doi.org/10.3389/fncom.2017.00082
Torres-Oviedo G, Ting LH (2010) Subject-specific muscle synergies in human balance control are consistent across different biomechanical contexts. J Neurophysiol 103(6):3084–3098. https://doi.org/10.1152/jn.00960.2009
Tresch MC, Saltiel P, Bizzi E (1999) The construction of movement by the spinal cord. Nat Neurosci 2:162–167
Tresch MC, Cheung VCK, d’Avella A (2006) Matrix factorization algorithms for the identification of muscle synergies: evaluation on simulated and experimental data sets. J Neurophysiol 95(4):2199–2212. https://doi.org/10.1152/jn.00222.2005
Turpin NA, Guevel A, Durand S, Hug F (2011) No evidence of expertise-related changes in muscle synergies during rowing. J Electromyogr Kinesiol 21(6):1030–1040. https://doi.org/10.1016/j.jelekin.2011.07.013
Turpin NA, Costes A, Moretto P, Watier B (2017) Can muscle coordination explain the advantage of using the standing position during intense cycling? J Sci Med Sport 20(6):611–616. https://doi.org/10.1016/j.jsams.2016.10.019
Turpin NA, Martinez R, Begon M (2020) Shoulder muscle activation strategies differ when lifting or lowering a load. Eur J Appl Physiol 120(11):2417–2429. https://doi.org/10.1007/s00421-020-04464-9
Valero-Cuevas FJ, Venkadesan M, Todorov E (2009) Structured variability of muscle activations supports the minimal intervention principle of motor control. J Neurophysiol 102(1):59–68. https://doi.org/10.1152/jn.90324.2008
Van Criekinge T, Vermeulen J, Wagemans K, Schroder J, Embrechts E, Truijen S, Hallemans A, Saeys W (2019) Lower limb muscle synergies during walking after stroke: a systematic review. Disabil Rehabil. https://doi.org/10.1080/09638288.2019.1578421
van der Krogt MM, Oudenhoven L, Buizer AI, Dallmeijer A, Dominici N, Harlaar J (2016) The effect of EMG processing choices on muscle synergies before and after BoNT-A treatment in cerebral palsy
Viviani P, Laissard G (1991) Timing control in motor sequences. In: Fagard J, Wolff PH (eds) Advances in Psychology, vol 81. North-Holland, Amsterdam, pp 1–36. doi: https://doi.org/10.1016/S0166-4115(08)60758-X
Wakeling JM, Horn T (2009) Neuromechanics of muscle synergies during cycling. J Neurophysiol 101(2):843–854. https://doi.org/10.1152/jn.90679.2008
Wong AL, Krakauer JW (2019) Why are sequence representations in primary motor cortex so elusive? Neuron 103(6):956–958. https://doi.org/10.1016/j.neuron.2019.09.011
Yang Z, Oja E (2010) Linear and nonlinear projective nonnegative matrix factorization. IEEE Trans Neural Netw 21(5):734–749
Yang JF, Winter DA (1984) Electromyographic amplitude normalization methods: improving their sensitivity as diagnostic tools in gait analysis. Arch Phys Med Rehabil 65(9):517–521
Yang Q, Logan D, Giszter SF (2019) Motor primitives are determined in early development and are then robustly conserved into adulthood. Proc Natl Acad Sci USA 116(24):12025–12034. https://doi.org/10.1073/pnas.1821455116
Zandvoort CS, van Dieen JH, Dominici N, Daffertshofer A (2019) The human sensorimotor cortex fosters muscle synergies through cortico-synergy coherence. Neuroimage 199:30–37. https://doi.org/10.1016/j.neuroimage.2019.05.041
Zelik KE, La Scaleia V, Ivanenko YP, Lacquaniti F (2014) Can modular strategies simplify neural control of multidirectional human locomotion? J Neurophysiol 111(8):1686–1702. https://doi.org/10.1152/jn.00776.2013
Zheng Z, Yang J, Zhu Y (2007) Initialization enhancer for non-negative matrix factorization. Eng Appl Artif Intell 20(1):101–110
Acknowledgements
The authors thank Benjie BARTOS for the English corrections. We also thank Tieme LARAQUE for his valuable comments on the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Michael Lindinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Turpin, N.A., Uriac, S. & Dalleau, G. How to improve the muscle synergy analysis methodology?. Eur J Appl Physiol 121, 1009–1025 (2021). https://doi.org/10.1007/s00421-021-04604-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-021-04604-9