Abstract
Purpose
To evaluate the efficacy of real-time PCR for 16S ribosomal DNA (16S r-DNA) and sequencing for diagnosing microbial keratitis.
Methods
We studied 272 eyes of 272 patients with keratitis. Eyes with keratitis were classified as “definite” (N = 118), “likely” (N = 71), or “non-bacterial” (N = 83) to have bacterial keratitis. The diagnostic efficacy of real-time PCR and conventional testing was determined by receiver operating characteristic analysis. The copy numbers of bacterial DNA and clinical characteristics were retrospectively analyzed for association with concordant culture results in the “definite” cases.
Results
The level of bacterial DNA was significantly associated with the diagnostic probability of the three diagnostic categories. The level of bacterial DNA had comparable diagnostic efficacy with the area under the curve (AUC) at 0.67, by culture at 0.65, and by smear testing at 0.73. The efficacy was significantly improved by combining the DNA level with the conventional culture testing with an AUC of 0.81. Analysis of the “definite” cases showed culture positivity in 51.8% (58 eyes), and of these, 41 eyes (70.7%) were higher than the cutoff PCR values and 40 eyes were identified by 16S r-DNA sequencing. In the culture-negative eyes, the level of bacterial DNA was significantly lower (P = 0.0008). Eyes with higher bacterial DNA levels had significantly concordant outcomes with sequencing and culture results (P = 0.006). Previous antibiotic treatments decreased the bacterial DNA amount by 0.09-fold, and it was a significant factor for discordance (P = 0.006).
Conclusion
Quantification of the bacterial DNA level and conventional testing improves the diagnostic efficacy of infectious bacterial keratitis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The diagnosis of infectious and non-infectious keratitis is difficult even for experienced clinicians. To assist in making the correct diagnosis, culture tests of corneal samples is the gold standard method with high specificity. However, this usually requires days or weeks to obtain the results, and often a report of no cultivatable bacteria is returned. When using smear tests, the results of gram staining are readily obtained; however, they do not identify the bacteria at the species level. In addition, it is often difficult to evaluate its relevance when only a few bacteria are observed.
Considering the limitations of these conventional testing methods, an examination of the bacterial genomic DNA has been used to supplement the laboratory-based methods. The 16S ribosomal DNA (16S r-DNA) gene of the bacteria genome is well conserved and has been tested for using broad-range PCR to detect bacteria in clinical specimens. Broad range bacterial PCR is especially advantageous in detecting slow-growing bacteria or pauci-bacteria such as those in the eyes.
In infectious eye diseases, broad-range bacterial PCR and sequencing have been reported to be efficacious for endophthalmitis and infectious keratitis, and they have been shown to have high efficacy in identifying the bacteria at the species level [1, 2]. However, this does not completely reflect the efficacy in an actual clinical setting, because the sensitivity and specificity improve when assessed in selected patients with presumed infection.
Currently, the quantitative nature of 16S r-DNA real-time PCR has not been appreciated for its efficacy. For example, infectious keratitis may have a higher level of 16S r-DNA copy numbers than non-infectious cases. Nevertheless, real-time PCR has not been become a routine clinical test performed on all patients suspected to have bacterial keratitis.
Thus, the purpose of this study was to determine whether the ability to obtain quantitative levels of 16S r-DNA by real-time PCR with sequencing will improve the diagnostic efficacy in non-selected patients with keratitis. For this, we analyzed non-selected (random), consecutive keratitis cases of infectious, non-infectious, and suspected keratitis using logistic regression analysis and receiver operating characteristic (ROC) analysis.
Materials and methods
Patients eligibility and diagnosis criteria of bacterial infection
Two hundred seventy-two patients with clinically diagnosed keratitis from 2009 to 2015 were retrospectively analyzed. One eye of each patient was analyzed. The mean age of the patients was 57.6 ± 22.7 years, and 136 patients (50.0%) were men. Corneal samples were collected from all, and broad-range quantitative real-time PCR was performed to determine the level of the 16S r-DNA.
The diagnosis of bacterial keratitis in eyes with a corneal ulcer was made based on the clinical characteristics [3, 4], responsiveness to appropriate antibiotics, and identification of the bacteria in the corneal scrapings by one or more of the following findings: (1) detection of bacteria in smear by Gram staining, (2) positive bacterial culture, and (3) detection of the specific pathogens, e.g., Staphylococcus aureus, Pseudomonas aeruginosa, and mycobacterium species, by conventional PCR.
The exclusion of bacterial keratitis and corneal ulcers as the diagnosis was based on the clinical characteristics of “non-bacterial” keratitis, lack of the clinical characteristic of bacterial infection, responsiveness to appropriate anti-fungal or anti-viral drugs, and differential diagnosis by the exclusion diagnostic criteria [3,4,5,6,7,8,9,10], e.g., identification of fungi or acanthamoeba cysts in the corneal scrapings stained with Fungiflora YⓇ, positive cultures of fungi or acanthamoeba, and the identification of a genome of fungi, acanthamoeba, human herpes virus, or adenovirus in the corneal scrapings by conventional PCR [11]. Cases with a final diagnosis of autoimmune keratitis or corneal ulcer were also classified as “non-bacterial” keratitis when these tests were negative.
Cases that did not meet the inclusion and exclusion criteria but were strongly suspected to have a bacterial infection were classified as “likely.” The final diagnosis was made after all the outcomes of the tests were obtained and patients were treated. To assure a definitive diagnosis of bacterial keratitis, three independent observers reviewed all of the medical records.
The diagnosis and classification of “definite,” “likely,” and “non-bacterial” were made by the three independent investigators who were masked to the 16S r-DNA findings. For confirmation of the classification, unsupervised program-based classification was used. For this, latent class analysis (LCA) was conducted to classify the disease categories. The diagnosis of definite bacterial keratitis was made when the diagnosis of the examiners agreed with the outcomes of the LCA.
The study protocol was approved by Tottori University Ethics Committee.
Broad-range quantitative real-time PCR
Samples at the first visit were collected from the corneal surface by gently touching it with an ophthalmic surgical sponge (Inami, Japan) and rinsing of the corneal surface with 400 μl of saline without touching the eyelids. The samples were stored frozen, and DNA was extracted within 6 days with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The 16S r-DNA of bacteria was amplified as described [5]. The number of 16S r-DNA copies was calculated using a standard curve generated by serial dilutions of cloned DNA fragments of 16S r-DNA [12]. The number of 16S r-DNA copies of the bacterial DNA was converted to logarithmic (log10) units, and it was adjusted by the logarithm of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) copy numbers [13].
Cloning and sequencing of 16S r-DNA
Cloned libraries of the 16S r-DNA were generated, and they were transformed into E. coli DH-5α competent cells. The transformed clones were plated, and colony PCR was performed to identify the colonies containing appropriately sized inserts. The insert-containing colonies were extracted for plasmid DNA and sequenced. The resulting sequences were used to identify the bacteria species using the basic local alignment search tool (BLAST). The identification of the genus or species was defined as accurate when there was a ≥97% or 99% homology, respectively [2].
Statistical analyses
Data are presented as the means ± standard deviations. Unpaired t test, Mann-Whitney U test, and ANOVA with post hoc analysis test were used to determine the significance of the differences among the groups. The 16S r-DNA was quantified in logarithmic copy numbers of the 16S r-DNA. Multivariate ordered logistic regression analysis was used to compute the odds ratio (OR) after adjustments for age and the log GAPDH units. The area under the curve (AUC) was calculated by the receiver operating characteristic (ROC) analysis and propensity score analysis. Statistical analyses were done with the Stata 15 software (Stata Corp, College Station, TX). A P value of < 0.05 was considered statistically significant.
Results
Among the 272 eyes of 272 patients, 118 eyes of 118 patients were diagnosed as “definite” bacterial keratitis, 71 eyes of 71 patients as “likely” bacterial keratitis, and 83 eyes of 83 patients as “non-bacterial” keratitis. Their mean age in these three groups was 57.6 ± 22.8 years, 62.0 ± 21.2 years, and 61.5 ± 18.7 years, respectively (P = 0.28; Table 1). The percentage of eyes of contact lens wearers was significantly higher in eyes in the definite bacterial keratitis group. No significant trend was observed in the percentage of eyes of diabetic patients or the use of antibiotics.
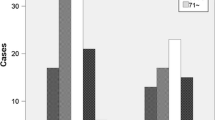
In the “definite” bacterial keratitis cases, the number of 16S r-DNA copies was 3.7 ± 2.0 log10 units which was significantly higher than that in the “non-bacterial” eyes at 2.5 ± 1.6 log10 units (P = 0.000, Fig. 1, top). For the “likely” bacterial keratitis eyes, an intermediate number of 2.9 ± 1.7 log10 units was found. The detection of bacteria by smear testing and culture had significantly higher positive findings in the “definite” cases, and the “likely” eyes had intermediate percentages of positive findings (Table 1).
Relationship between the bacterial 16S r-DNA copy number and the classification of bacterial keratitis. Top: The bacterial DNA copy number (16S r-DNA) of “definite” bacterial keratitis is significantly higher than in “non-bacterial” keratitis cases (P = 0.000). “Likely” cases have intermediate copy numbers of 16S r-DNA. Bottom: probability of predicting “definite,” “likely,” and “non-bacterial” keratitis depending on bacterial DNA copy numbers after GAPDH adjustment. A bacteria copy number higher than 103 has a significantly higher probability of “definite” bacterial keratitis classification
We assessed whether the patient characteristics, e.g., age, sex, diabetes, contact lens wear, and use of antibiotics on the initial visit, affected the outcome of each of the three examinations. Of these, the 16S r-DNA copy number and antibiotic pretreatment were significantly associated with the rate of culture positivity. For culture positivity, the OR of 16S r-DNA copy (log10 units) was 1.4 (95% CI 1.20–1.70, P = 0.000) and the OR for pretreatment with antibiotics was 0.31 (95% CI 0.17–0.57, P = 0.000 after age adjustment). These findings indicated that the high bacteria levels and no previous antibiotic use significantly increased the finding of culture positivity. No significant association was found in the diabetes and contact lens wearers.
Multiple linear regression was used to determine how the 16S r-DNA level was affected by the use of antibiotics. Previous antibiotic use significantly reduced the 16S r-DNA copy numbers to one-third the level of the eyes with no previous antibiotic use (31.8%, P = 0.04, after age adjustment).
Then, we assessed how each examination was associated with the degree of certainty of diagnosis, i.e., “definite,” “likely,” and “non-bacterial.” Ordered logistic regression analysis confirmed that the three bacterial examinations had highly significant associations with the degree of certainty. The 16S r-DNA with 5 log10 units had an OR of 5.09 for the degree of certainty. Smear testing had a high OR of 4.18, and the OR of culturing was 4.08 after adjustments for covariates including age, use of antibiotics, contact lens wear, and GAPDH (Table 2).
To understand how the use of 16S r-DNA can increase the probability of the degree of certainty depending on the copy numbers, the degree of diagnosis was calculated based on the logistic regression with covariate adjustments (Fig. 1, bottom). The probability of a “definite” diagnosis increased as the copy numbers increased, while that of “non-bacterial” diagnosis decreased. Both curves crossed at approximately 102 copies, and copy numbers above 103 copies favored a “definite” diagnosis of a bacterial infection.
Diagnostic efficacy is the important parameter for clinical application. Therefore, we assessed the diagnostic efficacy of the three bacterial examinations using ROC analysis (Fig. 2). The ROC curves were plotted for sensitivity and false-positive rates (1-specificity) of 16S r-DNA copies, bacteria culturing, and smear testing (Fig. 2). The area under the curve (AUC), which indicates the accuracy of the test, was 0.67 for the 16S r-DNA, 0.65 for the culturing, and 0.73 for the smear. The diagnostic efficacy of 16S r-DNA PCR was moderate as the AUC was not significantly different from the other two conventional testing methods. The 100% specificity for diagnosis was obtained when the copy number was more than 6.9 × 104 copies. This indicated that more than 6.9 × 104 copies favored a clinical diagnosis of “definite.”
Diagnostic efficacy of bacterial DNA real-time PCR and combined testing with conventional testing by receiver operating characteristic analysis. Top: diagnostic efficacy calculated by the area under the curve (AUC) of bacterial DNA real-time PCR (16S r-DNA: 16S) was 0.67, and it was not significantly different from culture and smear tests alone. Bottom: the AUC of combined testing of broad-range real-time PCR for 16S r-DNA, culture, and smear tests reached 0.81 which is significantly better than the results of single testing (top)
We then evaluated whether 16S r-DNA real-time PCR can be of further advantage for diagnosing when used in conjunction with the conventional testing. For this, the outcomes of testing were calculated by propensity score analysis, and they were assessed for improvements of the AUC compared with the conventional testing. Combined testing of bacterial culturing and 16S r-DNA PCR had an AUC of 0.72 which was significantly better than culture alone (P = 0.02). Combined testing of smear and 16S r-DNA also had a significantly higher AUC of 0.80 compared with the smear alone (P = 0.008). A combination of all three tests had a superior accuracy of 0.81 of AUC compared with culture testing (P = 0.0004). Thus, the addition of 16S r-DNA PCR significantly improved the diagnostic efficacy compared with each conventional testing alone.
To characterize the outcome of the 16S r-DNA PCR analyses, the optimal cutoff value was determined using the Youden index. For 16S r-DNA, the cutoff value was 1600 copies. When this cutoff value was used, 16S r-DNA PCR had a profile of specificity and positive predictive value of 67.5% and 73.5%, respectively (Table 3).
We also evaluated whether the copy numbers of 16S r-DNA will add further advantage to conventional culture testing. In the 112 “definite” cases that underwent culture testing together with 16S ribosomal PCR testing, 58 eyes (51.8%) were culture positive (Table 1). For the culture negative specimens, 2.9 ± 1.8 log10 units were detected, and the 16S r-DNA copy numbers was significantly lower than the culture positive eyes (P = 0.0008, Fig. 3, left). Because this difference suggested a significant number of bacteria were still undetected in the culture-negative cases, we examined whether 16S r-DNA sequencing will enhance the detection of the bacteria. We assessed the relationship between 16S r-DNA sequencing to culture positivity. Of the 112 “definite” cases, 58 eyes (51.8%) were culture positive. The culture-detected bacteria species were Staphylococcus aureus, Corynebacterium sp., coagulase-negative staphylococci (CNS), and Pseudomonas aeruginosa (Fig. 4, top). Staphylococcus aureus was the most frequently detected species.
Relationship between bacterial DNA copy number and culture-positivity or concordance with bacteria detection by culture and bacterial DNA sequencing in cases of “definite” bacterial keratitis. Left: bacterial DNA copy number in culture-positive eyes is significantly higher than that in culture-negative eyes. Right: comparison of sequencing of 16S r-DNA and culture detection of bacteria. Eyes with concordant outcomes had significantly higher bacterial DNA copy numbers than the discordant cases
Bacteria species detected in the “definite” bacterial keratitis cases by culturing and 16S r-DNA sequencing. Top: the culture detected bacteria were Staphylococcus aureus, Corynebacterium spp., and coagulase-negative staphylococci (CNS). Bottom: sequencing for “definite” cases detected different pathogenic species characterized by Moraxella spp., which are generally difficult to culture after antibiotics use
Of the 58 culture-positive eyes, 41 eyes (70.7%) were higher than the cutoff for PCR. Of the 54 culture-negative eyes, 29 eyes (53.7%) were higher than the cutoff. Samples with higher than the cutoff value were processed for sequence identification of the bacterial species. In the 41 culture-positive eyes, the species of the bacteria of 40 eyes (97.6%) was identified. In the 29 culture-negative eyes with higher than cutoff for PCR, the detection rate by sequencing was significantly lower, but 19 eyes (65.5%) were still identified (P = 0.000, Fisher test). The sequence-detected bacteria were uncultured bacteria followed by Moraxella spp., Proteobacterium, and Pseudomonas aeruginosa (Fig. 4, bottom).
Next, we assessed the degree of concordance of the identified bacterial species by culture and sequencing in the “definite” cases. When the culture-positive eyes in the “definite” cases were analyzed, 14 eyes (34.2%) matched the identified bacteria in the culture testing. Overall, the concordance rate of 70 “definite” cases including the culture negative cases was 20.0% (14 eyes). The mean 16S r-DNA copy number in the concordant eyes was significantly higher than that in the discordant eyes (P = 0.002, Fig. 3, right).
To determine under what circumstances the discordance occurred, we calculated the ORs of the testing outcomes and clinical characteristics that are associated with higher concordance rates. Logistic regression analysis showed that a strong positive association was observed for the copy numbers of 16S r-DNA (OR 2.85/per log10 units increase, 95% CI 1.35–6.01, P = 0.006, after age adjustment). In contrast, antibiotic pretreatment had a significant negative association (OR 0.06, 95% CI 0.009–0.45, P = 0.006, after age adjustment). Thus, in cases with high 16S r-DNA copy numbers detected, both tests were significantly matched. No specific bacterial species had significant associations with the culture and sequencing outcome (data not shown).
Finally, we calculated the effect of antibiotic use on the 16S r-DNA level using linear regression in all of the cases. Culture positivity was significantly associated with increased 16S r-DNA copy numbers by 58 folds (101.76, P = 0.000). Previous antibiotic use significantly affected the effect of culture positivity on the 16S r-DNA amount. A reduction of the 16S r-DNA amount by previous antibiotic use was calculated to be 0.09-fold (10–1.03 as interaction, P = 0.047).
Discussion
An important characteristic of 16S r-DNA real-time PCR is its high sensitive and quantitative nature. However, the quantitative aspects of 16S r-DNA analyses have been ignored, and it remains unclear how to interpret the discordant outcomes of culturing and PCR. We suggest that the quantitative evaluations of the PCR findings may improve the diagnostic efficacy and concordance. We found that high 16S r-DNA copy numbers indicated a high probability of “definite” bacterial keratitis and concordant outcomes with culture identification.
The usefulness of 16S r-DNA PCR in the diagnosis of infectious diseases has been investigated in different disciplines including ophthalmology. For example, it has been reported that 16S r-DNA PCR can be successfully used for early diagnosis of endophthalmitis or uveitis. It has been especially effective in examining samples from seemingly sterile sites or culture-negative samples. Bacterial evaluations using 16S r-DNA PCR have been successfully conducted for excised heart valves from suspected infectious endocarditis, blood samples from suspected sepsis cases, cerebrospinal samples from suspected meningitis cases, and bone or joint samples from suspected infection cases, and for the rapid detection of contamination of blood before transfusion [14].
The quantitative nature of 16S r-DNA has several important advantages. First, the quantification of the bacterial amount improved the accuracy of the diagnosis of bacterial infection, and higher copy numbers increased the probability of a bacterial infection. For example, a simple interpretation can be made for specimens with less than 1600 copies or over 6.9 × 104 copies (Fig. 1). Keratitis patients with over 6.9 × 104 copies were found in the “definite” cases. This allowed a prompt diagnosis and the initiation of appropriate antibiotics even when no bacteria were detected in the cultures. After an empirical treatment has commenced, the outcome of sequencing can be available within a week and the infecting microbe be identified at the species level.
Second, 16S r-DNA PCR analyses should be able to identify the bacteria in specimens collected after antibiotic treatment has commenced. Although antibiotic use is generally associated with culture negativity, 16S r-DNA PCR can still detect and identify the pathogenic bacteria. 16S r-RNA real-time PCR is especially useful for bacterial infections when cultures are negative. In our cohort, 35.2% of 54 culture negative cases were determined to be due to bacteria identified by sequencing. This ratio is comparable with 42.9% positivity from extensive analysis on non-selected systemic infection/ inflammation cases [15]. Analysis of infectious bacterial keratitis appears to provide comparable outcomes [16]. In contrast, two studies reported a much lower positivity in culture-negative cases [10, 17].
Some concern has been raised on whether amplified products represent actual pathogens [16] even though it has been shown that difficult-to-culture bacteria can cause keratitis [18]. For example, mycobacterium and spirochetes are not cultivatable or have very slow growth, and anaerobes, which constitute the majority of the proteobacteria, have very demanding culture conditions. All of these including very rare or environmental bacteria are more “likely” to have culture-negative results. However, it is possible that these organisms will be positive to 16S r-DNA real-time PCR.
A number of new bacteria have been recently identified by 16S r-DNA sequencing [19]. Schabereiter-Gurtner et al. showed that such under-appreciated bacteria can cause corneal ulcers in culture-negative cases [18]. Moreover, endophthalmitis has been shown by 16S r-DNA library sequencing to be caused by polymicrobial infections [20].
A combination of 16S r-DNA analysis with conventional bacterial examination had superior diagnostic efficacy as was shown by the ROC analysis (Fig. 2). This indicated that 16S r-DNA quantification improved the diagnostic efficacy by complementing the conventional tests which have unsatisfactory performance when tested individually.
To help the diagnosis of eye diseases, artificial intelligence (AI) programs have been gaining interest because of their effectiveness. We are currently developing the AI-based diagnosis algorithm using the clinical features and outcomes of examinations. By calculating the importance score, we have noted that the 16S r-DNA copy number was one of the most important features for the AI algorithm (data not shown). This further supports the idea that 16S r-DNA analysis is highly effective when combined with other clinical features.
Clinically, antibiotics are often started before bacterial examinations are complete and when an infection is suspected. This is one of the most significant causes of culture negativity, but PCR positivity can still be found as in the endocarditis cases after antibiotic treatment [15]. Using 394 systemic disease samples, Rampini showed that the use of antibiotics before specimen collection was a significant factor in reducing culture positivity [15]. Thus, specimen collection before antibiotic treatment was recommended. In our study of keratitis samples, we found that antibiotic use had a significant OR of 0.31 on culture testing which confirms its reduction of culture positivity. For 16S r-DNA PCR, the use of antibiotics reduced the copy number to 1/10; however, “definite” bacterial infection cases have high copy numbers, and such a reduction generally does not affect its positivity outcome. Thus, 16S r-DNA real-time PCR followed by sequencing appears better than culturing after the use of antibiotics.
Likely bacterial keratitis cases were complicated cases. These cases typically include culture/gram stain negative, but they are responsive to systemic or topical antibiotics without need of antivirals or antifungal drugs. This situation typically occurs when antibiotic treatment, which is an often insufficient dosing to be curative, was applied before the referral. Because an insufficient amount of antibiotics cleared bacteria on the corneal surface, the level of 16S r-DNA decreases when this category applies (Fig. 1b). However, sequencing of the samples can still identify bacterial species. This still provides valuable formation.
We did not detect a better performance of 16S r-DNA real-time PCR than conventional testing by ROC analysis. The limited efficacy of PCR as a single test was somewhat surprising because previous studies on body specimens indicated superior performance of PCR to culture tests especially for the specificity. The reported sensitivity and specificity sometimes reached 80–100% [15, 17]. The ocular surface including the cornea is considered to be a pauci-bacterial environment, but it is surrounded by the eyelid skin which harbors an abundant amount of microbial flora. This may explain why the quantification of microbial DNA alone had lower specificity than expected. The differences of the disease may be affected differently by the commensal bacteria in the different tissues.
The bacterial species detected by 16S r-DNA PCR sequencing were mostly concordant with the culture-detected bacterial species. In general, 16S r-DNA PCR and culturing had a concordance of 90.6% in infections other than ocular infections [14, 15]. In the ophthalmic literature, the outcome of bacteria detection in endophthalmitis is also concordant for 70–100% for culture-positive cases [2, 21, [22]. In contrast, bacterial keratitis samples appear to have a lower concordance of 63.2% [16] which is consistent with our outcome.
It is generally believed that endophthalmitis is caused by a single bacteria species; however, 16S r-DNA libraries have shown its polymicrobial nature [20]. Because the cornea and ocular surface are exposed to the environment, the involvement of the polymicrobial community may also be more relevant to infectious keratitis which might explain the lower concordance of PCR and culturing [14].
Another important issue that can affect the degree of concordance is antibiotic pretreatment which will decrease the culture positivity and the bacterial amount. For example, pathogenic Moraxella spp. are sensitive to antibiotics and readily become negative for culture testing. Indeed, we frequently detected the DNA of Moraxella spp. in culture-negative cases or Corynebacterium-detected cases by culturing. We found a discordance of culturing and PCR after antibiotic use. Thus, clinicians need to be aware that culture-detected bacteria after antibiotic use may not be the true causative pathogen.
Our findings indicated that the determination of the 16S r-DNA copy numbers is not the single most important test, but it is very helpful when combined with other conventional testing. In our tertiary referral hospital, 16S r-DNA testing has become the standard test for all the patients suspected for infectious keratitis. For patients who present without antibiotic pretreatment, we can assess the need for antibiotics as well as the strength of the required treatment based on the amount of 16S r-DNA. Higher levels of 16S r-DNA indicate the need of systemic antibiotics, while negative results indicate no or only prophylactic use of topical antibiotics.
There are several limitations in our study. Because our study design was retrospective, a selection bias may have affected our outcomes or variability. In addition, our facility is a tertiary referral hospital, and the studied populations may be biased toward cases that are refractory to treatment. As a single test, the variability of 16S r-DNA measurements is relatively large. When the eyes are infected with bacteria, 16S r-DNA drastically increases to more than 104 copies. However, the count becomes much lower when they are pretreated by antibiotics before referral. This relationship is similar to outcomes of culture or smear testing.
Our data indicated that outcomes of 16S r-DNA measurements were significantly associated with that of culture testing (OR 1.4, 95% CI 1.20–1.70; P = 0.000). The outcomes of 16S r-DNA measurements were significantly associated with the degree of certainty of diagnosis, “definite,” “likely,” and “non-bacterial” (P = 0.000). This strong association was similarly observed for culture testing as the gold standard (P = 0.000). These observations indicate that the estimated reliance of 16S r-DNA measurements is similar to that of culture testing although both tests have a very different meaning. Thus, the variability of 16S r-DNA results appears to be comparable with that of culture testing.
Although measurements of 16S r-DNA are sensitive, relying solely on 16S r-DNA yields insufficient diagnostic accuracy. However, integration of 16S r-DNA measurements, amplification, and sequencing with conventional tests significantly improve its usefulness and helps in deciding the treatment choices including the use of steroids (Fig. 2b).
We conclude that bacterial culture is still the gold standard test, and 16S ribosomal PCR is a good adjunctive test. We should remember that no single test can determine whether a lesion is infectious. However, we propose that the diagnostic performance of clinicians would be significantly improved by the use of 16S r-DNA real-time PCR and sequencing.
References
Sugita S, Ogawa M, Shimizu N, Morio T, Ohguro N, Nakai K, Maruyama K, Nagata K, Takeda A, Usui Y, Sonoda KH, Takeuchi M, Mochizuki M (2013) Use of a comprehensive polymerase chain reaction system for diagnosis of ocular infectious diseases. Ophthalmology 120:1761–1768. https://doi.org/10.1016/j.ophtha.2013.02.020
Chiquet C, Cornut PL, Benito Y, Thuret G, Maurin M, Lafontaine PO, Pechinot A, Palombi K, Lina G, Bron A, Denis P, Carricajo A, Creuzot C, Romanet JP, Vandenesch F (2008) Eubacterial PCR for bacterial detection and identification in 100 acute postcataract surgery endophthalmitis. Invest Ophthalmol Vis Sci 49:1971–1978
Chidambaram JD, Prajna NV, Larke NL, Palepu S, Lanjewar S, Shah M, Elakkiya S, Lalitha P, Carnt N, Vesaluoma MH, Mason M, Hau S, Burton MJ (2016) Prospective study of the diagnostic accuracy of the in vivo laser scanning confocal microscope for severe microbial keratitis. Ophthalmology 123:2285–2293. https://doi.org/10.1016/j.ophtha.2016.07.009
Bhadange Y, Das S, Kasav MK, Sahu SK, Sharma S (2015) Comparison of culture-negative and culture-positive microbial keratitis: cause of culture negativity, clinical features and final outcome. Br J Ophthalmol 99:1498–1502. https://doi.org/10.1136/bjophthalmol-2014-306414
Ikeda Y, Miyazaki D, Yakura K, Kawaguchi A, Ishikura R, Inoue Y, Mito T, Shiraishi A, Ohashi Y, Higaki S, Itahashi M, Fukuda M, Shimomura Y, Yagita K (2012) Assessment of real-time polymerase chain reaction detection of Acanthamoeba and prognosis determinants of Acanthamoeba keratitis. Ophthalmology 119:1111–1119
Ma JX, Wang LN, Zhou RX, Yu Y, Du TX (2016) Real-time polymerase chain reaction for the diagnosis of necrotizing herpes stromal keratitis. Int J Ophthalmol 9:682–686. https://doi.org/10.18240/ijo.2016.05.07
Kanavi MR, Javadi M, Yazdani S, Mirdehghanm S (2007) Sensitivity and specificity of confocal scan in the diagnosis of infectious keratitis. Cornea 26:782–786. https://doi.org/10.1097/ICO.0b013e318064582d
Hau SC, Dart JK, Vesaluoma M, Parmar DN, Claerhout I, Bibi K, Larkin DF (2010) Diagnostic accuracy of microbial keratitis with in vivo scanning laser confocal microscopy. Br J Ophthalmol 94:982–987. https://doi.org/10.1136/bjo.2009.175083
Dart JK, Saw VP, Kilvington S (2009) Acanthamoeba keratitis: diagnosis and treatment update 2009. Am J Ophthalmol 148:487–499 e482. https://doi.org/10.1016/j.ajo.2009.06.009
Panda A, Pal Singh T, Satpathy G, Wadhwani M, Monika M (2015) Comparison of polymerase chain reaction and standard microbiological techniques in presumed bacterial corneal ulcers. Int Ophthalmol 35:159–165. https://doi.org/10.1007/s10792-014-9925-9
Kakimaru-Hasegawa A, Kuo CH, Komatsu N, Komatsu K, Miyazaki D, Inoue Y (2008) Clinical application of real-time polymerase chain reaction for diagnosis of herpetic diseases of the anterior segment of the eye. Jpn J Ophthalmol 52:24–31. https://doi.org/10.1007/s10384-007-0485-7
Miyazaki D, Shimizu D, Shimizu Y, Inoue Y, Inoue T, Higaki S, Ueta M, Sugita S, PCRfocisg R-t (2018) Diagnostic efficacy of real-time PCR for ocular cytomegalovirus infections. Graefes Arch Clin Exp Ophthalmol 256:2413–2420. https://doi.org/10.1007/s00417-018-4111-9
Inata K, Miyazaki D, Uotani R, Shimizu D, Miyake A, Shimizu Y, Inoue Y (2018) Effectiveness of real-time PCR for diagnosis and prognosis of varicella-zoster virus keratitis. Jpn J Ophthalmol 62:425–431. https://doi.org/10.1007/s10384-018-0604-7
Maiwald M (2011) Broad-range PCR for detection and identification of bacteria. In: Persing DH, Tenover FC, Tang YW, Nolte FS, Hayden RT, van Belkum A (eds) Molecular microbilogy: diagnostic principles and practice. ASM Press, Washington DC, pp 491–505
Rampini SK, Bloemberg GV, Keller PM, Buchler AC, Dollenmaier G, Speck RF, Bottger EC (2011) Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis 53:1245–1251. https://doi.org/10.1093/cid/cir692
Kim E, Chidambaram JD, Srinivasan M, Lalitha P, Wee D, Lietman TM, Whitcher JP, Van Gelder RN (2008) Prospective comparison of microbial culture and polymerase chain reaction in the diagnosis of corneal ulcer. Am J Ophthalmol 146:714–723 723 e711
Fenollar F, Roux V, Stein A, Drancourt M, Raoult D (2006) Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 44:1018–1028. https://doi.org/10.1128/JCM.44.3.1018-1028.2006
Schabereiter-Gurtner C, Maca S, Kaminsky S, Rolleke S, Lubitz W, Barisani-Asenbauer T (2002) Investigation of an anaerobic microbial community associated with a corneal ulcer by denaturing gradient gel electrophoresis and 16S rDNA sequence analysis. Diagn Microbiol Infect Dis 43:193–199
Woo PC, Lau SK, Teng JL, Tse H, Yuen KY (2008) Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 14:908–934. https://doi.org/10.1111/j.1469-0691.2008.02070.x
Jayasudha R, Narendran V, Manikandan P, Prabagaran SR (2014) Identification of polybacterial communities in patients with postoperative, posttraumatic, and endogenous endophthalmitis through 16S rRNA gene libraries. J Clin Microbiol 52:1459–1466. https://doi.org/10.1128/JCM.02093-13
Lee AY, Akileswaran L, Tibbetts MD, Garg SJ, Van Gelder RN (2015) Identification of torque teno virus in culture-negative endophthalmitis by representational deep DNA sequencing. Ophthalmology 122:524–530. https://doi.org/10.1016/j.ophtha.2014.09.001
Sugita S, Shimizu N, Watanabe K, Katayama M, Horie S, Ogawa M, Takase H, Sugamoto Y, Mochizuki M (2011) Diagnosis of bacterial endophthalmitis by broad-range quantitative PCR. Br J Ophthalmol 95:345–349
Funding
This study was funded by Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Science, and Culture (15K20261).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Daisuke Shimizu declares that he has no conflict of interest.
Dai Miyazaki reports a speaker honorarium from Santen Pharmaceutical, a speaker honorarium from Senju Pharmaceutical, and a speaker honorarium from Alcon, outside the submitted work.
Fumie Ehara declares that she has no conflict of interest.
Yumiko Shimizu declares that she has no conflict of interest.
Ryu Uotani declares that he has no conflict of interest.
Koudai Inata declares that he has no conflict of interest.
Shin-ichi Sasaki has received research grants from the Japanese Ministry of Education, Science, and Culture (Grant-in-Aid 15K20261 for Scientific Research).
Yoshitsugu Inoue reports grants and a speaker honorarium from Senju Pharmaceutical Co., Ltd., grants from Santen Pharmaceutical Co., Ltd., and grants from Alcon Japan, Ltd., outside the submitted work.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shimizu, D., Miyazaki, D., Ehara, F. et al. Effectiveness of 16S ribosomal DNA real-time PCR and sequencing for diagnosing bacterial keratitis. Graefes Arch Clin Exp Ophthalmol 258, 157–166 (2020). https://doi.org/10.1007/s00417-019-04434-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-019-04434-8