Abstract
Whole exome sequencing (WES) is a recently developed technique in genetics research that attempts to identify causative mutations in complex, undiagnosed genetic conditions. Causative mutations are usually identified after filtering the hundreds of variants on WES from an individual’s DNA selected by the phenotype. We investigated a patient with a slowly progressive chronic axonal distal motor neuropathy and extrapyramidal syndrome using WES, in whom common genetic mutations had been excluded. Variant filtering identified potentially deleterious mutations in three known disease genes: DCTN1, KIF5A and NEFH, which have been all associated with similar clinical presentations of amyotrophic lateral sclerosis, Parkinsonism and/or hereditary spastic paraplegia. Predicting the functional effect of the mutations were analysed in parallel with detailed clinical investigations. This case highlights the difficulties and pitfalls of applying WES in patients with complex neurological diseases and serves as an instructive tale.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Whole exome sequencing (WES) is an increasingly available second-generation sequencing technique that identifies variants in the coding regions of the human genome [1]. In the field of neurology, it has helped patients by elucidating the cause of their often long-standing symptoms, and many previously undiagnosed patients with a range of neurological syndromes which can now be given the name of the causative gene and counselled appropriately. For researchers, WES has led to the rapid rise in the discovery of novel mutations in both known and novel disease genes, further unravelling the cellular function and relationships of various genes with human disease. However, one of the many limitations of WES is in its application to single patients with late-onset disease. WES of one individual’s DNA can identify up to 20,000 variants [1]. Even in patients with a characteristic clinical phenotype choosing the causative mutation remain a challenge if DNA from other affected or healthy relatives is not available to determine which variant segregates with the phenotype [2].
Here we describe a case wherein, we used WES to guide diagnosis in a patient with complex neurodegenerative disease.
Methods
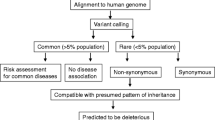
We assessed the patient’s clinical history, examination and serological, neurophysiological, radiological and histological investigations. Exome sequencing was carried out using genomic blood DNA by Illumina Truseq™ 62 Mb exome capture. We used our in-house bioinformatics pipeline to align data to the reference human genome (UCSC hg19), remove duplicate sequence reads (Picard v1.85) and variant detection (Varscan v2.29, Dindel v1.0110). Results were filtered for variants with a minor allele frequency less than 0.01 in several databases: dbSNP135, 1000 genomes (February 2012 data release), the National Heart, Lung and Blood Institute (NHLBI, NIH, Bethesda, MD) Exome Sequencing Project (ESP) 6500 exomes, and 238 unrelated in-house controls. Rare homozygous and compound heterozygous variants were defined, and protein altering and/or putative ‘disease-causing’ mutations, along with their functional annotation, were identified using ANNOVAR. Further filtering has been performed by gene ontology (GO) terms associated with neuronal function, and Online Mendelian Inheritance in Man (OMIM) disease descriptions related to neuropathy, ataxia or extrapyramidal disorders (i.e., the patient’s clinical phenotype). We carried out PCR (IMMOLASE™ DNA Polymerase, Bioline UK) and Sanger sequencing (BigDye® Terminator v3.1) of variants predicted to be deleterious by five online prediction tools (MutationTaster, SIFT, Polyphen2, A-GVGD, and LRT).
Results
Case report
A 74-year-old left-handed man originally presented at the age of 45 with right-sided back pain and sciatica as well as bilateral grip weakness. His gait had also changed but he was able to walk and play sports. Past medical history included poliomyelitis at the age of ten, during the 1950s epidemic.
Both of his parents died at age 88 and 72, and had no neuromuscular symptoms. He has one son and four daughters (28–36 years of age), and one daughter was recently diagnosed with multiple sclerosis. He has a 71-year-old healthy brother who was not available for testing.
At age 45, examination revealed wasting and weakness of the right sternocleidomastoid muscle, bilateral wasting of thenar and intrinsic hand muscles (Fig. 1a). In particular, the abductor pollicis brevis was weak bilaterally. There was distal lower limb weakness with foot drop but no pes cavus. Tone and reflexes were normal.
He was seen again at the age of 64 with worsening torticollis. He had jerky ocular pursuit movements and bilateral wasting of the tongue. His previous weakness had persisted with additional weakness in the shoulders. He had developed pes cavus bilaterally (Fig. 1a), brisk upper limb and knee reflexes with absent ankle reflexes bilaterally. He progressed to develop dysmetria and dysdiadochokinesis but no dysarthria.
More recent examination at 74 years revealed progression of the complex movement disorder with facial dystonia, head tremor, distal amyotrophy, ataxia and some rigidity at the wrists. He had a wide-based steppage gait with reduced arm swing.
Investigations
Basic haematological and biochemical investigations were normal except for gamma-glutamyl transferase (83 units/L, normal <70), creatine kinase (193 and 215 U/L, normal 10–190), random glucose (8.3 mmol/L), cholesterol (7.9 mmol/L, normal <5.2) and triglycerides (6.1 mmol/L, normal fasting level <1.8). Thyroid function tests, auto-antibodies, Vitamin E, 24-h urinary copper, alpha-fetoprotein, lysosomal enzymes, hexosaminidase A were all within normal limits.
Brain MRI revealed marked cerebellar atrophy and mild cortical atrophy without signal change (Fig. 1b). Cervical MRI showed mild spondylotic changes at C5/6 and C6/7. DatScan showed no abnormalities in dopamine metabolism. Nerve conduction studies demonstrated preserved sensory nerve action potentials, sensory and motor conduction velocities but symmetrically diminished compound motor potentials in all four limbs, suggesting that the syndrome could not be attributed to previous poliomyelitis. Electromyography revealed chronic neurogenic changes, suggesting spinal muscular atrophy or distal spinal muscular atrophy. With electromyography, unstable polyphasic units were observed with reduced interference and amplitudes up to 10 mV, confirming a chronic progressive anterior horn cell disease. Muscle biopsy showed only neurogenic changes.
Genetic analysis
Genetic tests performed before WES excluded Huntington’s disease (IT15), spinal muscular atrophy (SMN1), spinal bulbar muscular atrophy (AR), primary torsion dystonia (DYT1), and the common forms of spastic paraplegias (SPG4, REEP1, ATL1, SPG7). No mtDNA deletions were detected in his muscle DNA. Rare, potentially disease-causing variants found in filtered WES data are listed in Tables 1 and 2; we could confirm all variants in heterozygous state in the patient’s DNA by Sanger sequencing. No further mutations were detected in any of the known disease genes involved in inherited movement disorders, neuropathies or spastic paraplegias and no further family members were available for genetic analysis.
Discussion
We have found novel, potentially pathogenic mutations in the patient in multiple genes (DCTN1, KIF5A, NEFH) associated with various neurological syndromes. Based on previous reports, heterozygous mutations in all three genes can be associated with similar clinical presentation observed in our case. In further support of the pathogenic role, the selected variants have not been reported previously at all (KIF5A) or are extremely rare (one single heterozygous DCTN1 and NEFH variant in 65,000 exomes (http://exac.broadinstitute.org/).
DCTN1 mutations have been associated with a familial form of ALS (p.Gly59Ser) [3], familial progressive supranuclear palsy [4], Perry syndrome (p.Gly71Ala/Glu/Arg, p.Thr72Pro, and p.Gln74Pro) which is characterized by central hypoventilation and parkinsonism [5]. Our patient exhibited a distal motor neuropathy with ataxia and an extrapyramidal syndrome of tremor and focal dystonia—an inexact fit with the previously described phenotypes of DCTN1. However, the involvement of the spinal motor neurones in combination with a movement disorder resembles DCTN1-related disease and our patient may represent a new phenotype associated with DCTN1 mutation.
DCTN1 codes for the p150glued subunit of the dynactin complex, which together with dynein is responsible for retrograde transport of vesicle cargo in axons [6]. The C-terminal region of p150glued has been shown in N. crassa to be responsible for the interaction of the dynactin complex with membranous cargo [7]. However, previous work in HeLA and COS7 cells could not confirm that mutations in the C-terminal region of DCTN1 (Arg1101Lys and Thr1249Ile) are functionally pathogenic [8]. Our patient’s DCTN1 mutation is located to this region; nevertheless, this variant has not been reported previously and no functional data are available on it to date. However, functional in vivo imaging studies in patients possessing the variants associated with Perry syndrome, have demonstrated a reduced uptake of FDOPA, a marker of dopamine synthesis and storage [9].
Mutations in KIF5A are associated with spastic paraplegia (SPG10), and can also cause peripheral neuropathy, parkinsonism, retinitis pigmentosa and behavioural problems [10, 11]. Therefore, the pyramidal signs in our patient may also represent a KIF5A-related disease. KIF5A codes for the kinesin heavy chain which is responsible for anterograde transport of cargo (thought to be neurofilaments) towards the synapse [12]. The KIF5A mutation in our patient occurs in the neck region that links two domains in the protein [10]. To our knowledge, only one mutation (p. Ala361Val) has previously been reported in the neck region in spastic paraplegia [13]; however, its pathogenic role has not been confirmed in functional studies [14].
WES also detected a heterozygous 24-base pair deletion in the neurofilament heavy chain (NEFH) gene in our patient. Such deletions of a lysine-serine-proline unit, that shorten the repetitive tail region of the NEFH protein, have been associated with an increased risk of developing ALS [15], but no highly penetrant causative alleles have been described in this gene.
Another intriguing point is the poliomyelitis in our patient’s previous history. Whether the childhood neuropathy was indeed poliomyelitis, and coupled with a potentially deleterious mutation contributed to the development of a slowly progressive predominantly distal motor neuropathy, or the childhood onset polio-like neuropathy represents the first manifestation of a genetic condition remains unclear.
This case illustrates the problems that can be encountered in using next generation sequencing to guide the diagnosis of sporadic cases. Despite having confirmed potentially disease-causing mutations in DCTN1 and KIF5A, as well as a genetic predisposing variant in NEFH, we cannot surely prove that they are the cause for the patient’s phenotype without an informative family history, which is often not available in late-onset disease. The absence of clinical signs and symptoms in younger family members may simply reflect a later disease onset or low penetrance of the mutation rather than its non-pathogenicity. Moreover, we cannot exclude the possibility that more than one of the variants are relevant, and the different aspects of the patient’s condition potentially may have independent causes.
The case also highlights the importance of clinical history and examination in filtering through the enormous number of possible diagnoses generated by the results of next generation sequencing, and the need for functional studies at a molecular level to identify which mutation contributes to the cellular dysfunction. The overlapping phenotypes of genetic forms of ALS, HSP and dystonia highlight the importance of an unbiased testing approach, but also illustrate the difficulties of confirming the genetic diagnosis and functional tests provided evidence for the pathogenicity of these mutations are still lacking.
In conclusion, we present a patient with a complex neurological phenotype including predominantly distal motor neuropathy, extrapyramidal and cerebellar signs for whom we have identified a number of potentially pathogenic candidate mutations.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- HSP:
-
Hereditary spastic paraplegia
- CMT:
-
Charcot–Marie–Tooth disease
References
Bras J, Guerreiro R, Hardy J (2012) Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci 13(7):453–464
Yuan J, Higuchi Y, Nagado T, Nozuma S, Nakamura T, Matsuura E et al (2013) Novel mutation in the replication focus targeting sequence domain of DNMT1 causes hereditary sensory and autonomic neuropathy IE. J Peripher Nerv Syst 18(1):89–93
Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E et al (2003) Mutant dynactin in motor neuron disease. Nat Genet 33(4):455–456
Caroppo P, Le Ber I, Clot F et al (2014) DCtn1 mutation analysis in families with progressive supranuclear palsy—like phenotypes. JAMA Neurol 71(2):208–215
Wider C, Dachsel JC, Farrer MJ, Dickson DW, Tsuboi Y, Wszolek ZK (2010) Elucidating the genetics and pathology of Perry syndrome. J Neurol Sci 289(1–2):149–154
Moughamian AJ, Holzbaur ELF (2012) Dynactin is required for transport initiation from the distal axon. Neuron 74(2):331–343
Kumar S, Zhou Y, Plamann M (2001) Dynactin-membrane interaction is regulated by the C-terminal domains of p150Glued. EMBO Rep 2(10):939–944
Dixit R, Levy JR, Tokito M, Ligon LA, Holzbaur ELF (2008) Regulation of dynactin through the differential expression of p150Glued isoforms. J Biol Chem 283(48):33611–33619
Felicio AC, Dinelle K, Agarwal PA, McKenzie J, Heffernan N, Road JD et al (2014) In vivo dopaminergic and serotonergic dysfunction in DCTN1 gene mutation carriers. Mov Disord 29(9):1197–1201
Kawaguchi K (2013) Role of kinesin-1 in the pathogenesis of SPG10, a rare form of hereditary spastic paraplegia. Neurosci 19(4):336–344
Liu Y-T, Laurá M, Hersheson J, Horga A, Jaunmuktane Z, Brandner S et al (2014) Extended phenotypic spectrum of KIF5A mutations: From spastic paraplegia to axonal neuropathy. Neurology 83(7):612–619
Xia C-H, Roberts EA, Her L-S, Liu X, Williams DS, Cleveland DW et al (2003) Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol 161(1):55–66
LoGiudice M, Neri M, Falco M et al (2006) A missense mutation in the coiled-coil domain of the kif5a gene and late-onset hereditary spastic paraplegia. Arch Neurol 63(2):284–287
Ebbing B, Mann K, Starosta A, Jaud J, Schöls L, Schüle R et al (2008) Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet 17(9):1245–1252
Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C et al (1999) Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet 8(2):157–164
Acknowledgments
RH is supported by the Medical Research Council (UK) (G1000848) and the European Research Council (309548). PFC is a Wellcome Trust Senior Fellow in Clinical Science and National Institute for Health Research Senior Investigator. He receives funding from the Medical Research Council and the National Institute for Health Research Biomedical Research Centre for Ageing and Age-Related Disease award to the Newcastle upon Tyne Foundation Hospitals National Health Service Trust.
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical standard
Ethical approval has been obtained from local ethics committee and the patient gave written consent for the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Daud, D., Griffin, H., Douroudis, K. et al. Whole exome sequencing and the clinician: we need clinical skills and functional validation in variant filtering. J Neurol 262, 1673–1677 (2015). https://doi.org/10.1007/s00415-015-7755-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-015-7755-y