Abstract
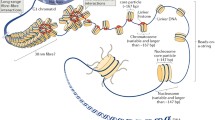
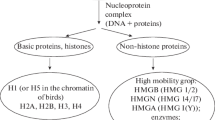
H1 linker histones are involved both in the maintenance of chromatin higher-order structure and in gene regulation. H1 binds to linker DNA regions on the surface of the nucleosome. In higher eukaryotes, H1 contains three distinct domains: a short N-terminal domain (NTD), a central globular domain, and a long C-terminal domain (CTD). Terminal domains determine subtype specificity and to a large extent the linker DNA binding and chromatin condensing properties of histone H1. This review is focused on the recent numerous studies that have provided insights in the role of H1 terminal domains in chromatin dynamics. The N- and C-terminal domains behave as intrinsically disordered proteins with coupled binding and folding. We examine the potential kinetic advantages of intrinsic disorder in the recognition of the specific H1 binding sites in chromatin. As typical intrinsically disordered regions, H1 terminal domains are post-translationally modified. Post-translational modifications in the NTD determine the interaction of histone H1 with other proteins involved in heterochromatin formation and transcriptional regulation, while phosphorylation by cyclin-dependent kinases modulates the secondary structure of the CTD and chromatin condensation. We review the arguments in favor of the involvement of H1 hyperphosphorylation in metaphase chromatin condensation and of partial phosphorylation in interphase chromatin relaxation. In addition, the interplay of histone H1 and other chromatin architectural proteins, such as proteins of the high-mobility group, protamines, and MeCP2, is associated with changes in chromatin structure.


Similar content being viewed by others
References
Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC (2007) Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. Journal of Biological Chemistry 282:15057–15064
An W, van Holde K, Zlatanova J (1998) The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. Journal of Biological Chemistry 273:26289–26291
Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proceedings of the National Academy of Sciences of the United States of America 95:14173–14178
Brown DT, Izard T, Misteli T (2006) Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nature Structural and Molecular Biology 13:250–255
Castaño J, Morera C, Sesé B, Boue S, Bonet-Costa C, Martí M, Roque A, Jordan A, Barrero MJ (2016) SETD7 regulates the differentiation of human embryonic stem cells. PLoS One 11:e0149502
Caterino TL, Fang H, Hayes JJ (2011) Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Molecular and Cellular Biology 31:2341–2348
Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M (2004) Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Molecular and Cellular Biology 24:4321–4328
Catez F, Ueda T, Bustin M (2006) Determinants of histone H1 mobility and chromatin binding in living cells. Nature Structural and Molecular Biology 13:305–310
Cato L, Stott K, Watson M, Thomas JO (2008) The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. Journal of Molecular Biology 384:1262–1272
Chu CS, Hsu PH, Lo PW, Scheer E, Tora L, Tsai HJ, Tsai MD, Juan LJ (2011) Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome. Journal of Biological Chemistry 286:35843–35851
Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE (2003) The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Molecular and Cellular Biology 23:8626–8636
Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R (2005) HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. Journal of Biological Chemistry 280:38090–38095
Doenecke D, Albig W, Bode C, Drabent B, Franke K, Gavenis K, Witt O (1997) Histones: genetic diversity and tissue-specific gene expression. Histochemistry and Cell Biology 107:1–10
Drabent B, Bode C, Doenecke D (1993) Structure and expression of the mouse testicular H1 histone gene (H1t). Biochimica et Biophysica Acta 1216:311–313
Eisert RJ, Kennedy SA, Waters ML (2015) Investigation of the β-sheet interactions between dHP1 chromodomain and histone 3. Biochemistry 54:2314–2322
Fang H, Clark DJ, Hayes JJ (2012) DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Research 40:1475–1484
Fischle WL, Franz H, Jacobs JA, Allis CD, Khorasanizadeh S (2008) Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. Journal of Biological Chemistry 283:19626–19635
Flanagan TW, Brown DT (2016) Molecular dynamics of histone H1. Biochimica et Biophysica Acta 1859:468–475
Flanagan TW, Files JK, Casano KR, George EM, Brown DT (2016) Photobleaching studies reveal that a single amino acid polymorphism is responsible for the differential binding affinities of linker histone subtypes H1.1 and H1.5. Biology Open. doi:10.1242/bio.016733
Ghosh RP, Nikitina T, Horowitz-Scherer RA, Gierasch LM, Uversky VN, Hite K, Hansen JC, Woodcock CL (2010) Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry 49:4395–410
Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T (2016) Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proceedings of the National Academy of Sciences of the United States of America 113:1238–1243
Gurley LR, Valdez JG, Buchanan JS (1995) Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. Journal of Biological Chemistry 270:27653–27660
Hansen JC, Lu X, Ross ED, Woody RW (2006) Intrinsic protein disorder, amino acid composition, and histone terminal domains. Journal of Biological Chemistry 281:1853–1856
Happel N, Stoldt S, Schmidt B, Doenecke D (2009) M phase-specific phosphorylation of histone H1.5 at threonine 10 by GSK-3. Journal of Molecular Biology 386:339–350
Hergeth SP, Dundr M, Tropberger P, Zee BM, Garcia BA, Daujat S, Schneider R (2011) Isoform-specific phosphorylation of human linker histone H1.4 in mitosis by the kinase Aurora B. Journal of Cell Science 124:1623–1628
Hutchinson JB, Cheema MS, Wang J, Missiaen K, Finn R, Gonzalez Romero R, Th’ng JP, Hendzel M, Ausió J (2015) Interaction of chromatin with a histone H1 containing swapped N- and C-terminal domains. Bioscience Reports 35(3)
Izzo A, Schneider R (2016) The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochimica et Biophysica Acta 1859:486–95
Izzo A, Kamieniarz-Gdula K, Ramírez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R (2013) The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Reports 3:2142–2154
Jacobs SA, Khorasanizadeh S (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080–2083
Kalashnikova AA, Winkler DD, McBryant SJ, Henderson RK, Herman JA, Deluca JG, Luger K, Prenni JE, Hansen JC (2013) Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Research 41:4026–4035
Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J, Scheer E, Tropel P, Wisniewski JR, Tora L, Viville S, Buettner R, Schneider R (2012) A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes and Development 26:797–802
Kowalski A, Pałyga J (2012) Linker histone subtypes and their allelic variants. Cell Biology International 36:981–996
Lennox RW, Cohen LH (1984) The alterations in H1 histone complement during mouse spermatogenesis and their significance for H1 subtype function. Developmental Biology 103:80–84
Lever MA, Th’ng JP, Sun X, Hendzel MJ (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873–876
Liao R, Mizzen CA (2016) Interphase H1 phosphorylation: regulation and functions in chromatin. Biochimica et Biophysica Acta 1859:476–85
Lopez R, Sarg B, Lindner H, Bartolomé S, Ponte I, Suau P, Roque A (2015) Linker histone partial phosphorylation: effects on secondary structure and chromatin condensation. Nucleic Acids Research 43:4463–4476
Lu X, Hansen JC (2004) Identification of specific functional subdomains within the linker histone H10 C-terminal domain. Journal of Biological Chemistry 279:8701–8707
Lu X, Hamkalo B, Parseghian MH, Hansen JC (2009) Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48:164–172
Luque A, Collepardo-Guevara R, Grigoryev S, Schlick T (2014) Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Research 42:7553–7560
Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I (2005) Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proceedings of the National Academy of Sciences of the United States of America 102:2808–2813
Mayor R, Izquierdo-Bouldstridge A, Millan-Arino L, Bustillos A, Sampaio C, Luque N, Jordan A (2015) Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. Journal of Biological Chemistry 290:7474–7491
McBryant SJ, Adams VH, Hansen JC (2006) Chromatin architectural proteins. Chromosome Research 14:39–51
Meyer S, Becker NB, Syed SH, Goutte-Gattat D, Shukla MS, Hayes JJ, Angelov D, Bednar J, Dimitrov S, Everaers R (2011) From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nucleic Acids Research 39:9139–9154
Millán-Ariño L, Izquierdo-Bouldstridge A, Jordan A (2016) Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochimica et Biophysica Acta 1859:510–519
Misteli T, Gunjan A, Hock R, Bustin M, Brown DT (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877–881
Nightingale K, Dimitrov S, Reeves R, Wolffe AP (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO Journal 15:548–561
Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL (2007) MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. Journal of Biological Chemistry 282:28237–28245
Öberg C, Belikov S (2012) The N-terminal domain determines the affinity and specificity of H1 binding to chromatin. Biochemical and Biophysical Research Communications 420:321–324
Parseghian MH (2015) What is the role of histone H1 heterogeneity? A functional model emerges from a 50 year mystery. AIMS Biophysics 2:724–772
Ponte I, Vidal-Taboada JM, Suau P (1998) Evolution of the vertebrate H1 histone class: evidence for the functional differentiation of the subtypes. Molecular and Biological Evolution 15:702–708
Postnikov Y, Bustin M (2010) Regulation of chromatin structure and function by HMGN proteins. Biochimica et Biophysica Acta 1799:62–68
Postnikov YV, Bustin M (2016) Functional interplay between histone H1 and HMG proteins in chromatin. Biochimica et Biophysica Acta 1859:462–467
Raghuram N, Carrero G, Th’ng J, Hendzel MJ (2009) Molecular dynamics of histone H1. Biochemistry and Cell Biology 87:189–206
Raghuram N, Strickfaden H, McDonald D, Williams K, Fang H, Mizzen C, Hayes JJ, Th’ng JP, Hendzel MJ (2013) Pin1 promotes histone H1 dephosphorylation and stabilizes its binding to chromatin. Journal of Cell Biology 203:57–71
Roque A, Iloro I, Ponte I, Arrondo JL, Suau P (2005) DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. Journal of Biological Chemistry 280:32141–32147
Roque A, Ponte I, Suau P (2007) Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophysical Journal 93:2170–2177
Roque A, Ponte I, Arrondo JL, Suau P (2008) Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Research 36:4719–4726
Roque A, Ponte I, Suau P (2009) Role of charge neutralization in the folding of the carboxy-terminal domain of histone H1. Journal of Physical Chemistry B 113:12061–12066
Roque A, Ponte I, Suau P (2016) Interplay between histone H1 structure and function. Biochimica et Biophysica Acta 1859:444–454
Roth SY, Allis CD (1992) Chromatin condensation: does histone H1 dephosphorylation play a role? Trends in Biochemical Sciences 17:93–98
Saitoh Y, Laemmli UK (1994) Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609–622
Sarg B, Helliger W, Talasz H, Förg B, Lindner HH (2006) Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. Journal of Biological Chemistry 281:6573–6580
Sarg B, Chwatal S, Talasz H, Lindner HH (2009) Testis-specific linker histone H1t is multiply phosphorylated during spermatogenesis. Identification of phosphorylation sites. Journal of Biological Chemistry 284:3610–3618
Sassone-Corsi P (2002) Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296:2176–2178
Shintomi K, Takahashi TS, Hirano T (2015) Reconstitution of mitotic chromatids with a minimum set of purified factors. Nature Cell Biology 17:1014–1023
Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Molecular Cell 37:457–468
Stasevich TJ, Mueller F, Brown DT, McNally JG (2010) Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO Journal 29:1225–1234
Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S (2010) Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proceedings of the National Academy of Sciences of the United States of America 107:9620–9625
Talasz H, Helliger W, Puschendorf B, Lindner H (1996) In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry 35:1761–1767
Talasz H, Sarg B, Lindner H (2009) Site-specifically phosphorylated forms of H1.5 and H1.2 localized at distinct regions of the nucleus are related to different processes during the cell cycle. Chromosoma 118:693–709
Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, Cross GA, Cui L, Dimitrov SI, Doenecke D, Eirin-López JM, Gorovsky MA, Hake SB, Hamkalo BA, Holec S, Jacobsen SE, Kamieniarz K, Khochbin S, Ladurner AG, Landsman D, Latham JA, Loppin B, Malik HS, Marzluff WF, Pehrson JR, Postberg J, Schneider R, Singh MB, Smith MM, Thompson E, Torres-Padilla ME, Tremethick DJ, Turner BM, Waterborg JH, Wollmann H, Yelagandula R, Zhu B, Henikoff S (2012) A unified phylogeny-based nomenclature for histone variants. Epigenetics & Chromatin 5:7
Terme JM, Millán-Ariño L, Mayor R, Luque N, Izquierdo-Bouldstridge A, Bustillos A, Sampaio C, Canes J, Font I, Sima N, Sancho M, Torrente L, Forcales S, Roque A, Suau P, Jordan A (2014) Dynamics and dispensability of variant-specific histone H1 Lys-26/Ser-27 and Thr-165 post-translational modifications. FEBS Letters 588:2353–2362
Th’ng JP, Guo XW, Swank RA, Crissman HA, Bradbury EM (1994) Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. Journal of Biological Chemistry 269:9568–9573
Th’ng JP, Sung R, Ye M, Hendzel MJ (2005) H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. Journal of Biological Chemistry 280:27809–27814
Trieschmann L, Postnikov YV, Rickers A, Bustin M (1995) Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Molecular and Cellular Biology 15:6663–6669
Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D (2009) Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the jumonji domain-containing JMJD2/KDM4 proteins. Journal of Biological Chemistry 284:8395–8405
Uversky VN (2013) A decade and a half of protein intrinsic disorder, biology still waits for physics. Protein Sciences 22:693–724
Vicent GP, Wright RH, Beato M (2016) Linker histones in hormonal gene regulation. Biochimica et Biophysica Acta 1859:520–525
Vila R, Ponte I, Collado M, Arrondo JL, Jiménez MA, Rico M, Suau P (2001) DNA-induced alpha-helical structure in the NH2-terminal domain of histone H1. Journal of Biological Chemistry 276:46429–46435
Vila R, Ponte I, Jimenez MA, Rico M, Suau P (2002) An induciblé helix-Gly-Gly-helix motif in the N-terminal domain of histone H1e: a CD and NMR study. Protein Sciences 11:214–220
Vyas P, Brown DT (2012) N- and C-terminal domains determine differential nucleosomal binding geometry and affinity of linker histone isotypes H1(0) and H1c. Journal of Biological Chemistry 287:11778–11787
Wang X, Moore SC, Laszckzak M, Ausió J (2000) Acetylation increases the alpha-helical content of the histone tails of the nucleosome. Journal of Biological Chemistry 275:35013–35020
Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, Dundr M, Garcia BA, Daujat S, Schneider R (2010) Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics & Chromatin 3:7
Wright RH, Castellano G, Bonet J, Le Dily F, Font-Mateu J, Ballaré C, Nacht AS, Soronellas D, Oliva B, Beato M (2012) CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes and Development 26:1972–1983
Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN (2007) Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. Journal of Proteome Research 6:1917–1932
Yan W, Ma L, Burns KH, Matzuk MM (2003) HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proceedings of the National Academy of Sciences of the United States of America 100:10546–10551
Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, Nardulli AM, Hager GL, Kelleher NL, Mizzen CA (2010) Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. Journal of Cell Biology 189:407–415
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported in part by the Ministerio de Ciencia e Innovación (BFU2008–00460).
Conflict of interest
The authors declare that they have no competing interest.
Animal experiments
This article does not contain any studies with human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Roque, A., Ponte, I. & Suau, P. Post-translational modifications of the intrinsically disordered terminal domains of histone H1: effects on secondary structure and chromatin dynamics. Chromosoma 126, 83–91 (2017). https://doi.org/10.1007/s00412-016-0591-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-016-0591-8