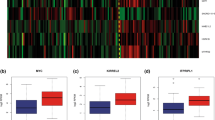
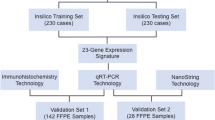
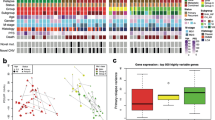
Abstract
Medulloblastoma (MB), one of the most common malignant pediatric brain tumor, is a heterogenous disease comprised of four distinct molecular groups (WNT, SHH, Group 3, Group 4). Each of these groups can be further subdivided into second-generation MB (SGS MB) molecular subgroups, each with distinct genetic and clinical characteristics. For instance, non-WNT/non-SHH MB (Group 3/4) can be subdivided molecularly into eight distinct and clinically relevant tumor subgroups. A further molecular stratification/summarization of these SGS MB would allow for the assignment of patients to risk-associated treatment protocols. Here, we performed DNA- and RNA-based analysis of 574 non-WNT/non-SHH MB and analyzed the clinical significance of various molecular patterns within the entire cohort and the eight SGS MB, with the aim to develop an optimal risk stratification of these tumors. Multigene analysis disclosed several survival-associated genes highly specific for each molecular subgroup within this non-WNT/non-SHH MB cohort with minimal inter-subgroup overlap. These subgroup-specific and prognostically relevant genes were associated with pathways that could underlie SGS MB clinical-molecular diversity and tumor-driving mechanisms. By combining survival-associated genes within each SGS MB, distinct metagene sets being appropriate for their optimal risk stratification were identified. Defined subgroup-specific metagene sets were independent variables in the multivariate models generated for each SGS MB and their prognostic value was confirmed in a completely non-overlapping validation cohort of non-WNT/non-SHH MB (n = 377). In summary, the current results indicate that the integration of transcriptome data in risk stratification models may improve outcome prediction for each non-WNT/non-SHH SGS MB. Identified subgroup-specific gene expression signatures could be relevant for clinical implementation and survival-associated metagene sets could be adopted for further SGS MB risk stratification. Future studies should aim at validating the prognostic role of these transcriptome-based SGS MB subtypes in prospective clinical trials.


Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Amayiri N, Swaidan M, Ibrahimi A, Hirmas N, Musharbash A, Bouffet E et al (2021) Molecular subgroup is the strongest predictor of medulloblastoma outcome in a resource-limited country. JCO Glob Oncol 7:1442–1453. https://doi.org/10.1200/GO.21.00127
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737–754. https://doi.org/10.1016/j.ccell.2017.05.005
Chatterjee A, Maitre M, Dasgupta A, Sridhar E, Gupta T (2022) Multidisciplinary management of medulloblastoma: consensus, challenges, and controversies. Methods Mol Biol 2423:215–235. https://doi.org/10.1007/978-1-0716-1952-0_19
Cruzeiro GAV, Salomão KB Jr, de Biagi CAO, Baumgartner M, Sturm D, Lira RCP (2019) A simplified approach using Taqman low-density array for medulloblastoma subgrouping. Acta Neuropathol Commun 7:33. https://doi.org/10.1186/s40478-019-0681-y
D’Arcy CE, Nobre LF, Arnaldo A, Ramaswamy V, Taylor MD, Naz-Hazrati L et al (2020) Immunohistochemical and nanostring-based subgrouping of clinical medulloblastoma samples. J Neuropathol Exp Neurol 79:437–447. https://doi.org/10.1093/jnen/nlaa005
das Chagas PF, de Sousa GR, Veronez LC, Martins-da-Silva A, Corrêa CAP, Cruzeiro GAV et al (2022) Identification of ITPR1 as a hub gene of group 3 medulloblastoma and coregulated genes with potential prognostic values. J Mol Neurosci 72:633–641. https://doi.org/10.1007/s12031-021-01942-3
Delaidelli A, Dunham C, Santi M, Negri GL, Triscott J, Zheludkova O et al (2022) Clinically tractable outcome prediction of Non-WNT/Non-SHH medulloblastoma based on TPD52 IHC in a multicohort study. Clin Cancer Res 28:116–128. https://doi.org/10.1158/1078-0432.CCR-21-2057
Gershanov S, Madiwale S, Feinberg-Gorenshtein G, Vainer I, Nehushtan T, Michowiz S et al (2021) Classifying medulloblastoma subgroups based on small, clinically achievable gene sets. Front Oncol 11:637482. https://doi.org/10.3389/fonc.2021.637482
Goschzik T, Schwalbe EC, Hicks D, Smith A, Zur Muehlen A, Figarella-Branger D et al (2018) Prognostic effect of whole chromosomal aberration signatures in standard-risk, non-WNT/non-SHH medulloblastoma: a retrospective, molecular analysis of the HIT-SIOP PNET 4 trial. Lancet Oncol 19:1602–1616. https://doi.org/10.1016/S1470-2045(18)30532-1
Hovestadt V, Ayrault O, Swartling FJ, Robinson GW, Pfister SM, Northcott PA (2020) Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat Rev Cancer 20(1):42–56. https://doi.org/10.1038/s41568-019-0223-8
Kaur K, Jha P, Pathak P, Suri V, Sharma MC, Garg A et al (2019) Approach to molecular subgrouping of medulloblastomas: comparison of NanoString nCounter assay versus combination of immunohistochemistry and fluorescence in-situ hybridization in resource constrained centres. J Neurooncol 143:393–403. https://doi.org/10.1007/s11060-019-03187-y
Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA et al (2012) Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123:473–484. https://doi.org/10.1007/s00401-012-0958-8
Korshunov A, Okonechnikov K, Stichel D, Schrimpf D, Delaidelli A, Tonn S et al (2022) Gene expression profiling of Group 3 medulloblastomas defines a clinically tractable stratification based on KIRREL2 expression. Acta Neuropathol 144:339–352. https://doi.org/10.1007/s00401-022-02460-1
Luo Z, Dong X, Yu J, Xia Y, Berry KP, Rao R et al (2021) Genomic and transcriptomic analyses reveals ZNF124 as a critical regulator in highly aggressive medulloblastomas. Front Cell Dev Biol 9:634056. https://doi.org/10.3389/fcell.2021.634056
Mutlu M, Tekin C, Ak Aksoy S, Taskapilioglu MO, Kaya S, Balcin RN et al (2021) Long non-coding RNAs as predictive markers of group 3 medulloblastomas. Neurol Res 17:1–10. https://doi.org/10.1080/01616412.2021.1975223
Mynarek M, Obrecht D, Sill M, Sturm D, Kloth-Stachnau K, Selt F et al (2023) Identification of low and very high-risk patients with non-WNT/non-SHH medulloblastoma by improved clinico-molecular stratification of the HIT2000 and I-HIT-MED cohorts. Acta Neuropathol 145:97–112. https://doi.org/10.1007/s00401-022-02522-4
Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T et al (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317. https://doi.org/10.1038/nature22973
Okonechnikov K, Federico A, Schrimpf D, Sievers P, Sahm F, Koster J et al (2023) Comparison of transcriptome profiles between medulloblastoma primary and recurrent tumors uncovers novel variance effects in relapses. Acta Neuropathol Commun 11(1):7. https://doi.org/10.1186/s40478-023-01504-1
Pfister SM, Reyes-Múgica M, Chan JKC, Hasle H, Lazar AJ, Rossi S et al (2022) A summary of the inaugural WHO classification of pediatric tumors: transitioning from the optical into the molecular era. Cancer Discov 12:331–355. https://doi.org/10.1158/2159-8290.CD-21-1094
Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J et al (2009) Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 27:1627–1636. https://doi.org/10.1200/JCO.2008.17.9432
Qin C, Pan Y, Li Y, Li Y, Long W, Liu Q (2021) Novel molecular hallmarks of Group 3 medulloblastoma by single-cell transcriptomics. Front Oncol 11:622430. https://doi.org/10.3389/fonc.2021.622430
Ramaswamy V, Remke M, Bouffet E, Bailey S, Clifford SC, Doz F et al (2016) Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol 31:821–831. https://doi.org/10.1007/s00401-016-1569-6
Riemondy KA, Venkataraman S, Willard N, Nellan A, Sanford B, Griesinger AM et al (2022) Neoplastic and immune single cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma. Neuro Oncol 24:273–276. https://doi.org/10.1093/neuonc/noab135
Schwalbe EC, Lindsey JC, Nakjang S, Crosier S, Smith AJ, Hicks D et al (2017) Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 18:958–971. https://doi.org/10.1016/S1470-2045(17)30243-7
Sharma T, Schwalbe EC, Williamson D, Sill M, Hovestadt V, Mynarek M et al (2019) Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol 138:309–326. https://doi.org/10.1007/s00401-019-02020-0
Shih DJ, Northcott PA, Remke M, Korshunov A, Ramaswamy V, Kool M et al (2014) Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol 32:886–896. https://doi.org/10.1200/JCO.2013.50.9539
Shuangshoti S, Tadadontip P, Techavichit P, Thorner PS, Shuangshoti S, Teerapakpinyo C (2020) Simplified molecular subtyping of medulloblastoma for reduced cost and improved turnaround time. Appl Immunohistochem Mol Morphol 28:538–543. https://doi.org/10.1097/PAI.0000000000000794
Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC et al (2012) Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123:465–472. https://doi.org/10.1007/s00401-011-0922-z
Thompson EM, Keir ST, Venkatraman T, Lascola C, Yeom KW, Nixon AB et al (2017) The role of angiogenesis in Group 3 medulloblastoma pathogenesis and survival. Neuro Oncol 19:1217–1227. https://doi.org/10.1093/neuonc/nox033
Williamson D, Schwalbe EC, Hicks D, Aldinger KA, Lindsey JC, Crosier S et al (2022) Medulloblastoma group 3 and 4 tumors comprise a clinically and biologically significant expression continuum reflecting human cerebellar development. Cell Rep 40:111162. https://doi.org/10.1016/j.celrep.2022.111162
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Korshunov, A., Okonechnikov, K., Schrimpf, D. et al. Transcriptome analysis stratifies second-generation non-WNT/non-SHH medulloblastoma subgroups into clinically tractable subtypes. Acta Neuropathol 145, 829–842 (2023). https://doi.org/10.1007/s00401-023-02575-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-023-02575-z