Abstract
Amyotrophic lateral sclerosis is characterized by progressive loss of motor neurons in the brain and spinal cord. Mutations in several genes, including FUS, TDP43, Matrin 3, hnRNPA2 and other RNA-binding proteins, have been linked to ALS pathology. Recently, Pur-alpha, a DNA/RNA-binding protein was found to bind to C9orf72 repeat expansions and could possibly play a role in the pathogenesis of ALS. When overexpressed, Pur-alpha mitigates toxicities associated with Fragile X tumor ataxia syndrome (FXTAS) and C9orf72 repeat expansion diseases in Drosophila and mammalian cell culture models. However, the function of Pur-alpha in regulating ALS pathogenesis has not been fully understood. We identified Pur-alpha as a novel component of cytoplasmic stress granules (SGs) in ALS patient cells carrying disease-causing mutations in FUS. When cells were challenged with stress, we observed that Pur-alpha co-localized with mutant FUS in ALS patient cells and became trapped in constitutive SGs. We also found that FUS physically interacted with Pur-alpha in mammalian neuronal cells. Interestingly, shRNA-mediated knock down of endogenous Pur-alpha significantly reduced formation of cytoplasmic stress granules in mammalian cells suggesting that Pur-alpha is essential for the formation of SGs. Furthermore, ectopic expression of Pur-alpha blocked cytoplasmic mislocalization of mutant FUS and strongly suppressed toxicity associated with mutant FUS expression in primary motor neurons. Our data emphasizes the importance of stress granules in ALS pathogenesis and identifies Pur-alpha as a novel regulator of SG dynamics.







Similar content being viewed by others
References
Anderson P, Kedersha N (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10:430–436. doi:10.1038/nrm2694
Anderson P, Kedersha N, Ivanov P (2015) Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849:861–870. doi:10.1016/j.bbagrm.2014.11.009
Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA et al (2013) ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA 110:E736–E745. doi:10.1073/pnas.1222809110
Ash PE, Vanderweyde TE, Youmans KL, Apicco DJ, Wolozin B (2014) Pathological stress granules in Alzheimer’s disease. Brain Res 1584:52–58. doi:10.1016/j.brainres.2014.05.052
Ayala YM, Pagani F, Baralle FE (2006) TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett 580:1339–1344. doi:10.1016/j.febslet.2006.01.052
Barmada SJ, Ju S, Arjun A, Batarse A, Archbold HC, Peisach D, Li X, Zhang Y, Tank EM, Qiu H et al (2015) Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci USA 112:7821–7826. doi:10.1073/pnas.1509744112
Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S (2010) Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci 30:639–649. doi:10.1523/JNEUROSCI.4988-09.2010
Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ, Quaresma AJ, Nickerson JA, Bosco DA (2013) Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener 8:30. doi:10.1186/1750-1326-8-30
Belzil VV, Gendron TF, Petrucelli L (2013) RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci 56:406–419. doi:10.1016/j.mcn.2012.12.006
Belzil VV, St-Onge J, Daoud H, Desjarlais A, Bouchard JP, Dupre N, Camu W, Dion PA, Rouleau GA (2011) Identification of a FUS splicing mutation in a large family with amyotrophic lateral sclerosis. J Hum Genet 56:247–249. doi:10.1038/jhg.2010.162
Bergemann AD, Johnson EM (1992) The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol 12:1257–1265
Bergemann AD, Ma ZW, Johnson EM (1992) Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol 12:5673–5682
Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ Jr, Sapp P, McKenna-Yasek D, Brown RH Jr, Hayward LJ (2010) Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19:4160–4175. doi:10.1093/hmg/ddq335
Boyd JD, Lee-Armandt JP, Feiler MS, Zaarur N, Liu M, Kraemer B, Concannon JB, Ebata A, Wolozin B, Glicksman MA (2014) A high-content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J Biomol Screen 19:44–56. doi:10.1177/1087057113501553
Buchan JR (2014) mRNP granules. Assembly, function, and connections with disease. RNA Biol 11:1019–1030. doi:10.4161/15476286.2014.972208
Buchan JR, Kolaitis RM, Taylor JP, Parker R (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153:1461–1474. doi:10.1016/j.cell.2013.05.037
Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276:36337–36343. doi:10.1074/jbc.M104236200
Buratti E, Baralle FE (2010) The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol 7:420–429 (pii:12205)
Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem 280:37572–37584. doi:10.1074/jbc.M505557200
Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20:1774–1784. doi:10.1093/emboj/20.7.1774
Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A (2012) TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem 287:15635–15647. doi:10.1074/jbc.M111.333450
Cooper-Knock J, Bury JJ, Heath PR, Wyles M, Higginbottom A, Gelsthorpe C, Highley JR, Hautbergue G, Rattray M, Kirby J et al (2015) C9ORF72 GGGGCC expanded repeats produce splicing dysregulation which correlates with disease severity in amyotrophic lateral sclerosis. PLoS ONE 10:e0127376. doi:10.1371/journal.pone.0127376
Cziko AM, McCann CT, Howlett IC, Barbee SA, Duncan RP, Luedemann R, Zarnescu D, Zinsmaier KE, Parker RR, Ramaswami M (2009) Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics 182:1051–1060. doi:10.1534/genetics.109.103234
D’Alton S, Altshuler M, Lewis J (2015) Studies of alternative isoforms provide insight into TDP-43 autoregulation and pathogenesis. RNA 21:1419–1432. doi:10.1261/rna.047647.114
Da CS, Cleveland DW (2011) Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol 21:904–919. doi:10.1016/j.conb.2011.05.029
Daigle JG, Lanson NA, Jr., Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB (2013) RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. doi:10.1093/hmg/dds526
Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G (2012) TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res 1462:16–25. doi:10.1016/j.brainres.2012.02.032
Di Salvio M, Piccinni V, Gerbino V, Mantoni F, Camerini S, Lenzi J, Rosa A, Chellini L, Loreni F, Carri MT et al (2015) Pur-alpha functionally interacts with FUS carrying ALS-associated mutations. Cell Death Dis 6:e1943. doi:10.1038/cddis.2015.295
Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I (2014) An ALS-associated mutation in the FUS 3′-UTR disrupts a microRNA-FUS regulatory circuitry. Nat Commun 5:4335. doi:10.1038/ncomms5335
Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B et al (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 29:2841–2857. doi:10.1038/emboj.2010.143
Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD (2014) Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci 34:8083–8097. doi:10.1523/JNEUROSCI.0543-14.2014
Finelli MJ, Liu KX, Wu Y, Oliver PL, Davies KE (2015) Oxr1 improves pathogenic cellular features of ALS-associated FUS and TDP-43 mutations. Hum Mol Genet 24:3529–3544. doi:10.1093/hmg/ddv104
Freibaum BD, Chitta RK, High AA, Taylor JP (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res 9:1104–1120. doi:10.1021/pr901076y
Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15:5383–5398. doi:10.1091/mbc.E04-08-0715
Hackman P, Sarparanta J, Lehtinen S, Vihola A, Evila A, Jonson PH, Luque H, Kere J, Screen M, Chinnery PF et al (2013) Welander distal myopathy is caused by a mutation in the RNA-binding protein TIA1. Ann Neurol 73:500–509. doi:10.1002/ana.23831
Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M, DDD study, Magee AC, Turnpenny PD, Baralle D (2014) Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet 51:806–813. doi:10.1136/jmedgenet-2014-102798
Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST (2007) Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55:556–564. doi:10.1016/j.neuron.2007.07.020
Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST (2003) RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39:739–747
Johnson EM (2003) The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res 23:2093–2100
Johnson EM, Chen PL, Krachmarov CP, Barr SM, Kanovsky M, Ma ZW, Lee WH (1995) Association of human Pur alpha with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Pur alpha recognition element. J Biol Chem 270:24352–24360
Johnson EM, Daniel DC, Gordon J (2013) The pur protein family: genetic and structural features in development and disease. J Cell Physiol 228:930–937. doi:10.1002/jcp.24237
Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J (2006) Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res 83:929–943. doi:10.1002/jnr.20806
Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442
Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y et al (2003) Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol 23:6857–6875
Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A et al (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495:467–473. doi:10.1038/nature11922
Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, Bonini NM (2014) Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 46:152–160. doi:10.1038/ng.2853
King OD, Gitler AD, Shorter J (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462:61–80. doi:10.1016/j.brainres.2012.01.016
Klar J, Sobol M, Melberg A, Mabert K, Ameur A, Johansson AC, Feuk L, Entesarian M, Orlen H, Casar-Borota O et al (2013) Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat 34:572–577. doi:10.1002/humu.22282
Kwiatkowski TJ Jr, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T et al (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323:1205–1208. doi:10.1126/science.1166066
Lagier-Tourenne C, Polymenidou M, Cleveland DW (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19:R46–R64. doi:10.1093/hmg/ddq137
Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C et al (2012) Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 15:1488–1497. doi:10.1038/nn.3230
Lalani SR, Zhang J, Schaaf CP, Brown CW, Magoulas P, Tsai AC, El-Gharbawy A, Wierenga KJ, Bartholomew D, Fong CT et al (2014) Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet 95:579–583. doi:10.1016/j.ajhg.2014.09.014
Lancaster AK, Nutter-Upham A, Lindquist S, King OD (2014) PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30:2501–2502. doi:10.1093/bioinformatics/btu310
Lanson NA Jr, Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB (2011) A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet 20:2510–2523. doi:10.1093/hmg/ddr150
Lanson NA Jr, Pandey UB (2012) FUS-related proteinopathies: lessons from animal models. Brain Res. doi:10.1016/j.brainres.2012.01.039
Liu H, Johnson EM (2002) Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res 30:2417–2426
Lloyd RE (2013) Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip Rev RNA 4:317–331. doi:10.1002/wrna.1162
Matus S, Bosco DA, Hetz C (2014) Autophagy meets fused in sarcoma-positive stress granules. Neurobiol Aging 35:2832–2835. doi:10.1016/j.neurobiolaging.2014.08.019
Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I (2012) FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J 31:4502–4510. doi:10.1038/emboj.2012.319
Nishimoto Y, Ito D, Yagi T, Nihei Y, Tsunoda Y, Suzuki N (2010) Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J Biol Chem 285:608–619. doi:10.1074/jbc.M109.022012
Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K (2002) Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem 277:37804–37810. doi:10.1074/jbc.M203608200
Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D (2012) Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep 13:759–764. doi:10.1038/embor.2012.90
Polymenidou M, Lagier-Tourenne C, Hutt KR, Bennett CF, Cleveland DW, Yeo GW (2012) Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res 1462:3–15. doi:10.1016/j.brainres.2012.02.059
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C et al (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14:459–468. doi:10.1038/nn.2779
Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, Barmada SJ, Finkbeiner S, Lui H, Carlton CE et al (2014) ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest 124:981–999. doi:10.1172/JCI72723
Ramaswami M, Taylor JP, Parker R (2013) Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154:727–736. doi:10.1016/j.cell.2013.07.038
Renton AE, Chio A, Traynor BJ (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23. doi:10.1038/nn.3584
Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA (2014) Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging 35:2822–2831. doi:10.1016/j.neurobiolaging.2014.07.026
Scaramuzzino C, Casci I, Parodi S, Lievens PM, Polanco MJ, Milioto C, Chivet M, Monaghan J, Mishra A, Badders N et al (2015) Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85:88–100. doi:10.1016/j.neuron.2014.12.031
Shorter J, Taylor JP (2013) Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis 1:e25200. doi:10.4161/rdis.25200
Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P et al (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323:1208–1211. doi:10.1126/science.1165942
Vance C, Scotter EL, Nishimura AL, Troakes C, Mitchell JC, Kathe C, Urwin H, Manser C, Miller CC, Hortobagyi T et al (2013) ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet 22:2676–2688. doi:10.1093/hmg/ddt117
Vanderweyde T, Youmans K, Liu-Yesucevitz L, Wolozin B (2013) Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59:524–533. doi:10.1159/000354170
White MK, Johnson EM, Khalili K (2009) Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle 8:1–7
Wolozin B (2014) Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med 17:47–52
Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS et al (2013) Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA 110:7778–7783. doi:10.1073/pnas.1219643110
Zeng LH, Okamura K, Tanaka H, Miki N, Kuo CH (2005) Concomitant translocation of Puralpha with its binding proteins (PurBPs) from nuclei to cytoplasm during neuronal development. Neurosci Res 51:105–109. doi:10.1016/j.neures.2004.09.009
Author information
Authors and Affiliations
Corresponding author
Additional information
J G. Daigle and K. Krishnamurthy contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2015_1530_MOESM1_ESM.tif
Supplementary material 1 Fig. 1: Quantification of Pur-alpha and FUS positive SGs. (a). ALS-Patient lymphoblastoid cells carrying FUS R518G and age/sex matched population control were stress with 0.5 mM sodium arsenite for 1 h 30 min and stained for endogenous Pur-alpha (anti-PURA), G3 PB, and DAPI. The number of cells containing SGs was counted and the number of cells containing pur-alpha positive SGs was quantified. Percentage of cells colocalized with both pur-alpha and G3BP under stress conditions was determined. Bar graph represents the average from three separate fields. Approximately, 80 % of cells contained G3BP-positive SGs (b). Control, ALS patient cells carrying FUS R518G, and FUS R521C mutations were stained with anti-FUS N-terminal and anti-G3BP. Number of cells containing FUS incorporated into SGs was divided by the total number of cells containing SGs to give a percent of cells with FUS positive SGs. N = ~200 cells. ANOVA with Tukey’s posthoc analysis was applied. (**P = 0.01). (TIFF 10113 kb)
401_2015_1530_MOESM2_ESM.tif
Supplementary material 2 Fig. 2: Pur-alpha localizes to SGs in mammalian neuronal cells. (a). Neuroblastoma cells (N2A) were stained with Dapi (blue), anti-HA7 (green), and anti-G3BP (red). In unstressed conditions, G3BP was diffuse in the cytoplasm. (b). Under stress (0.5 mM sodium arsenite), G3BP decorated cytoplasmic stress granules. (c). N2A cells transiently transfected with HA-Pur-alpha. Pur-alpha was localized to the cytoplasm. Pur-alpha overexpression did not induce G3BP-positive SGs under unstressed conditions. (d). N2A cells transiently expressing HA-Pur-alpha under stress conditions (0.5 mM sodium arsenite) displayed Pur-alpha in cytoplasmic puncta that colocalized with G3BP in the cytoplasm. Arrowheads indicate cytoplasmic SGs. Arrows indicate SGs containing HA-Pur-alpha. (TIFF 33410 kb)
401_2015_1530_MOESM3_ESM.tif
Supplementary material 3 Fig. 3: Pur-alpha co-localizes with SGs in primary motor neurons. Primary motor neurons were cultured in normal medium conditions (unstressed) and in medium conditions containing 0.5 mM sodium arsenite for 90 min. Under unstressed conditions endogenous Pur-alpha (green) and G3BP (red) appeared diffuse in the cytoplasm. MAP2 chicken antibody (a neuronal marker) was used to label the neuronal processes. Under stress conditions, Pur-alpha was present in large cytoplasmic puncta which colocalized with SG marker G3BP (depicted as large yellow puncta in the merge image). Arrows indicate Pur-alpha incorporation into SGs. (TIFF 11896 kb)
401_2015_1530_MOESM4_ESM.tif
Supplementary material 4 Fig. 4: Pur-alpha gets incorporated into SGs under heat stress. Neuroblastoma cells (N2A) untransfected, transfected with HA-Pur-alpha WT, and TET-inducible FUS-R521C-GFP cells were cultured in DMEM at 37 °C on Poly-D Lysine coated coverslips (unstressed). Duplicate groups were cultured in DMEM at 42 °C for 3 h 30 min (stressed) then fixed. Immunoflourescense was performed with anti-G3BP (red), anti-HA7 to illuminate HA-Pur-alpha expressing cells (green), and FUS R521C-GFP expressing cells (green). In the Pur-alpha expressing cells colocalization of Pur-alpha (green) and G3BP positive SGs (red) = yellow puncta, in FUS-R521C-GFP (green) and G3BP positive SGs (red) = yellow granules. SGs are indicated with arrows with tails. SGs containing Pur-alpha or FUS are pointed out with arrowheads. (TIFF 41170 kb)
401_2015_1530_MOESM5_ESM.tif
Supplementary material 5 Fig. 5: Pur-alpha is a component of in SGs formed under H 2 0 2 stress. Neuroblastoma cells (N2A) untransfected, transfected with HA-Pur-alpha WT, and TET-inducible FUS-R521C-GFP cells were cultured in DMEM at 37 °C (unstressed). Duplicate groups were cultured in 1 mM H2O2 in DMEM for 1 h 30 min then fixed. Immunofluorescence was performed with anti-G3BP (red), anti-HA7 to illuminate HA-Pur-alpha expressing cells (green), and FUS R521C-GFP expressing cells (green). In the Pur-alpha expressing cells colocalization of Pur-alpha (green) and G3BP positive SGs (red) = yellow puncta, in FUS-R521C-GFP (green) and G3BP positive SGs (red) = yellow granules. SGs are indicated with arrows with tails. SGs containing Pur-alpha or FUS are pointed out with arrowheads. (TIFF 39456 kb)
401_2015_1530_MOESM6_ESM.tif
Supplementary material 6 Fig. 6: Pur-alpha is trapped in SGs containing ALS- linked FUS R521C. Mammalian neuroblastoma cells (N2a) stably expressing TET inducible GFP-FUS R521C were plated on poly-D lysine coverslips then treated with 300 ng of doxycycline (DOX) for 24 h to induce expression of FUS. DOX was included in control un-transfected cells and was present in the medium throughout the experiment. Both groups were treated with 0.5 mM sodium arsenite for 90 min to induce SGs. After SGs were formed, cells washed with DMEM and allowed to recover in DMEM + DOX for 90 min at 37 °C. Following the recovery time period Pur-alpha (red) and G3BP (white) in control cells appeared diffuse with few small puncta remaining in the cytoplasm. In cells expressing GFP FUS (green), Pur-alpha (red) remains in G3BP positive SGs that were not resolve when stress was removed (Depicted as Yellow/white puncta in the cytoplasm). Unresolved SGs showing co-localization of FUS, Pur-alpha, and G3BP are indicated with arrows. (TIFF 17169 kb)
401_2015_1530_MOESM7_ESM.tif
Supplementary material 7 Fig. 7: Pur-alpha shRNA knockdown blocks the formation of TIAR-positive stress granules. HEK293T cells expressing Pur-alpha shRNA-GFP were stressed with 0.5 mM sodium arsenite for 1.5 h. Cells were stained with anti-TIAR (Red). Cytoplasmic SGs can be seen in adjacent untransfected cells but are absent in cells expressing Pur-alpha shRNA- GFP (green). (TIFF 31674 kb)
401_2015_1530_MOESM8_ESM.tif
Supplementary material 8 Fig. 8: Pur-alpha shRNA knockdown reduces the mRNA levels of SG components (a) qPCR was performed using primers specific to endogenous Pur-alpha as well as GAPDH. (b) qPCR was performed using primers specific to endogenous FMR1 as well as GAPDH. (c) qPCR was performed using primers specific to endogenous G3BP1 as well as GAPDH. (d) qPCR was performed using primers specific to endogenous TIAL1 as well as GAPDH. Relative mRNA levels were quantified and normalized to GAPDH. Graph shows fold changes in mRNA expression. ANOVA was performed with Tukey’s post hoc analysis. Error bars represent ± SEM. n.s = not significant. (*P < 0.05, **P = 0.01, ***P = 0.001). (TIFF 8819 kb)
401_2015_1530_MOESM9_ESM.tif
Supplementary material 9 Fig. 9: Pur-alpha shRNA knockdown inhibits the formation of SGs under heat stress. HEK293T cells expressing Pur-alpha shRNA –GFP were stressed with 42 °C for 4 h. Cells were fixed then stained with Anti-G3BP (Red) and Dapi (blue). Cytoplasmic SGs in neighboring untransfected cells are indicated with arrows. (TIFF 16643 kb)
401_2015_1530_MOESM10_ESM.tif
Supplementary material 10 Fig. 10: Pur-alpha knockdown in HEK293T cells does not affect G3BP protein levels. (a). HEK293T cells stably expressing pur-alpa shRNA were used to generate total lysates ~ 35,000 cells per well. Western blot analysis reveals a G3BP positive band at ~ 70 kDa. (b) Band quantification was taken from three separate blots and normalized to tubulin loading control. Average band intensities were analyzed with ANOVA revealing no significant differences across groups. (TIFF 13212 kb)
401_2015_1530_MOESM11_ESM.tif
Supplementary material 11 Fig. 11: Pur-alpha shRNA knockdown does not affect the levels of endogenous FUS mRNA and protein levels. HEK293T cells stably expressing Pur-alpha shRNA–GFP constructs. (a). Total lysates were made from 35,000 cells and resolved by SDS PAGE. Western blots were probed with anti-FUS (N-term) and anti-tubulin. FUS protein levels were unaltered in Pur-alpha shRNA samples compared to the scrambled shRNA control. (b). qPCR was performed using primers specific to endogenous FUS as well as GAPDH. Relative mRNA levels were quantified and normalized to GAPDH. Graph shows fold changes in FUS mRNA levels. ANOVA was performed with Tukey’s post hoc analysis. Error bars represent ± SEM. n.s = not significant. (TIFF 13212 kb)
401_2015_1530_MOESM12_ESM.tif
Supplementary material 12 Fig. 12: Pur-alpha shRNA does not influence formation of p-bodies under stress conditions. HEK293T cells expressing Pur-alpha shRNA constructs were stressed with 0.5 mM sodium arsenite then labeled with anti-GW182 [p-body marker]. (a) Immunofluorescence revealed p-bodies were present in control cells expressing scrambled shRNA as well as in cells expressing Pur-alpha shRNA. (b) Quantification indicates that there was no difference in number of p-bodies per cell in Pur-alpha shRNA groups compared to controls. ANOVA was performed with Tukey’s post hoc multiple comparison analysis. The graph shows the average number of p-bodies per cell. n = 300 per/group. (n.s = not significant) error bars represent SEM. Arrows indicate p-bodies counted in the quantification. (TIFF 31936 kb)
401_2015_1530_MOESM13_ESM.tif
Supplementary material 13 Fig. 13: Pur-alpha shRNA knockdown does not affect endogenous FUS distribution. HEK293T cells expressing Pur-alpha shRNA were labeled for endogenous hFUS [anti-FUS-N-terminal]. FUS (red) was primarily located in the nucleus in control cells expressing scrambled shRNA. In cells expressing Pur-alpha shRNA-GFP (green), the localization did not change. (TIFF 27587 kb)
401_2015_1530_MOESM14_ESM.tif
Supplementary material 14 Fig. 14: Pur-alpha over-expression promotes the formation of SG at earlier time points compared to control. Neuroblastoma cells, Control, expressing HA-Pur-alpha, and expressing FUS R521C-GFP were treated with 0.5 mM sodium arsenite. Time-lapse induction of SGs was conducted with treatments stopped after 10 min, 20 min, 30 min, 45, mins, and 1 h. Fixed cells were stained with anti-G3BP (red). In untreated conditions (time 0), G3BP appeared diffuse in the cytoplasm. SG (G3BP positive granules) began to form at 20 min time-point in control and FUS R521C conditions. In Pur-alpha over expression SG appear to form as early as 10 min an earlier time point compared to control. (TIFF 27720 kb)
401_2015_1530_MOESM15_ESM.tif
Supplementary material 15 Fig. 15: FUS R521C expression stalls SG disassembly and over-expression of pur-alpha accelerates SG turnover. Neuroblastoma cells (untransfected control, HA-Pur-alpha transient expression, and FUS R521C-GFP inducible expression) were stressed with 0.5 mM sodium arsenite for 1 h. Stress treatment was removed, washed with DMEM, then replaced with fresh DMEM. Groups were then allowed to recover for 2 h 30 min at 37 °C. (a). Schematic depicts the formation of SGs over time. Cells treated with 0.5 mM sodium arsenite for 1 h showed a robust induction of SGs. Once the stress is removed (replaced with fresh DMEM) SGs begin do dissociate. After 2 h and 30 min SGs have dissolved in control cells. (b) Immunoflourescense shows control cells with disassembled (diffuse red staining in the cytoplasm) SGs and in some cells SG still remaining (red puncta). HA-Pur-alpha expressing cells (green) SGs have dissolved. Cells containing FUS R521C (green) FUS is incorporated into SG (yellow) and these granules do not dissociate after 2 h 30 min post stress. Co-localization is indicated with arrows. (c). At 60× magnification, Z-projections at three separate fields were imaged. Number cells containing > 1 SG were quantified and divided by the total number of DAPI positive cells to obtain percentage of cells containing SGs. ANOVA was performed with Tukey’s multiple comparisons post hoc analysis. N = ~ 250 (Error bars represent ± SEM. (*P < 0.05, **P = 0.01). (TIFF 41168 kb)
401_2015_1530_MOESM16_ESM.tif
Supplementary material 16 Fig. 16: Identification of the prion-like domain in Pur-alpha. Using a predictive algorithm [54], which has been used to screen other RNA binding proteins for their prion-like properties, we analyzed the Pur-alpha sequence for the presence of prion like domain (s) [48, 54]. We used the full-length amino acid sequence of human Pur-alpha to determine whether the protein contains functional domains with prion-like properties. For comparison, we show that a large portion of the FUS protein is predicted to possess prion-like properties. (TIFF 8125 kb)
401_2015_1530_MOESM17_ESM.tif
Supplementary material 17 Fig. 17: Primary motor neurons expressing FUS R521G have significantly smaller soma compared to control. Mouse primary motor neurons exogenously expressing FUS R521G, FUS WT, FUS WT + Pur-alpha, and FUS R521G + Pur-alpha were measured using ImageJ. Horizontal and vertical dimensions were measured to quantify the neuron soma size. Student t test was performed between groups. Error bars represent ± SEM (*P < 0.05). (TIFF 5067 kb)
401_2015_1530_MOESM18_ESM.tif
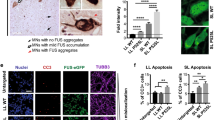
Supplementary material 18 Fig. 18: Expression of Pur-alpha reduces total number of SGs as well as FUS-positive SGs. (a) Representative confocal images of FUS R521G alone (top panel) and Pur-alpha co-expressing neurons (bottom panel) labeled for G3BP1 after sodium arsenite stress. Co-localization of G3BP1 with mutant FUS is shown by white arrows. Scale = 5 µm (b). SG numbers were significantly reduced in cells co-expressing Pur-alpha (* P = 0.039) (c) FUS positive SGs are significantly less in the cells co-expressing Pur-alpha compared to mutant FUS cells (*P0.034). N = 9. (TIFF 1175 kb)
Rights and permissions
About this article
Cite this article
Daigle, J.G., Krishnamurthy, K., Ramesh, N. et al. Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity. Acta Neuropathol 131, 605–620 (2016). https://doi.org/10.1007/s00401-015-1530-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-015-1530-0