Abstract
Introduction
Neural tube defects (NTDs) are a group of common and severe congenital birth defects that occur during early embryonic development due to incomplete closure of the neural tube. The genetic architecture of human NTDs, including spina bifida and hydrocephalus, is highly heterogeneous, with multiple genes/loci and both gene-gene and gene-environment interactions involved. Hence, the variation in outcomes also most likely relates to a combination of the severity of different variants in multiple genes and genetic modifiers affecting the biochemical traits.
Methods
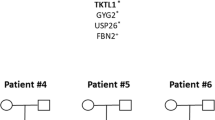
Here, we present a multiple-spouse family with one pedigree lineage where three brothers are affected with NTDs—two lumbar spina bifidas without hydrocephalus and one obstructive hydrocephalus. We sequenced the exomes of three NTD patients and their parents.
Results
The analysis revealed a heterozygous c.844ins68 variant in CBS, which was carried by all affected individuals and inherited from their mother. All affected individuals had a variable set of additional low frequency deleterious variants in PTK7, PLCD4, IL4I1 or RASSF4 as likely causal loci contributing to the disease development.
Conclusion
This report extends the current knowledge of the genetic background of NTDs and proposes that common and low frequency variants in genes involved mostly in one-carbon metabolism or planar cell polarity (PCP) pathways can act in an additive manner to increase the genetic risk of the disease.

Similar content being viewed by others
References
Greene NDE, Copp AJ (2014) Neural tube defects. Annu Rev Neurosci 37:221–242
Kibar Z, Capra V, Gros P (2007) Toward understanding the genetic basis of neural tube defects. Clin Genet 71:295–310
Detrait ER, George TM, Etchevers HC, Gilbert JR, Vekemans M, Speer MC (2005) Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol Teratol 27(3):515–524
Greene NDE, Stanier P, Copp AJ (2009) Genetics of human neural tube defects. Hum Mol Genet 18(R2):R113–R129
Harris MJ, Juriloff DM (2007) Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth defects res. A Clin Mol Teratol 79(3):187–210
Krupp DR, Soldano KL, Garrett ME, Cope H, Ashley-Koch AE, Gregory SG (2014) Missing genetic risk in neural tube defects: can exome sequencing yield an insight? Birth Defects Res A Clin Mol Teratol 100(8):642–646
Lemay P, Guyot MC, Tremblay É, Dionne-Laporte A, Spiegelman D, Henrion É, Diallo O, De Marco P, Merello E, Massicotte C, Désilets V, Michaud JL, Rouleau GA, Capra V, Kibar Z (2015) Loss-of-function de novo mutations play an important role in severe human neural tube defects. J Med Genet 52(7):493–497
Bassuk AG, Kibar Z (2009) Genetic basis of neural tube defects. Semin Pediatr Neurol 16:101–110
Andrew J Copp, Nicholas DE Greene, (2009) Genetics and development of neural tube defects. The Journal of Pathology:n/a-n/a
Sponholz C, Kramer M, Schöneweck F, Menzel U, Inanloo Rahatloo K, Giamarellos-Bourboulis EJ, Papavassileiou V, Lymberopoulou K, Pavlaki M, Koutelidakis I, Perdios I, Scherag A, Bauer M, Platzer M, Huse K (2016) Polymorphisms of cystathionine beta-synthase gene are associated with susceptibility to sepsis. Eur J Hum Genet 24(7):1041–1048
De Franchis R, Botto LD, Sebastio G, Ricci R, Iolascon A, Capra V, Andria G, Mastroiacovo P (2002) Spina bifida and folate-related genes: a study of gene-gene interactions. Genet Med 4:126–130
Rubini M, Brusati R, Garattini G, Magnani C, Liviero F, Bianchi F, Tarantino E, Massei A, Pollastri S, Carturan S, Amadori A, Bertagnin E, Cavallaro A, Fabiano A, Franchella A, Calzolari E (2005) Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate. Am J Med Genet 136A:368–372
Ouyang S, Liu Z, Li Y, Ma F, Wu J (2014) Cystathionine beta-synthase 844ins68 polymorphism is unrelated to susceptibility to neural tube defects. Gene 535(2):119–123
Nazki FH, Sameer AS, Ganaie BA (2014) Folate: metabolism, genes, polymorphisms and the associated diseases. Gene 533(1):11–20
Yaliwal LV, Desai RM (2012) Methylenetetrahydrofolate reductase mutations, a genetic cause for familial recurrent neural tube defects. Indian J Hum Genet 18(1):122–124
Shaw GM, Lu W, Zhu H, Yang W, Briggs FBS, Carmichael SL, Barcellos LF, Lammer EJ, Finnell RH (2009) 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Med Genet 10:49
Imbard A, Benoist J-F, Blom HJ (2013) Neural tube defects, folic acid and methylation. Int J Environ Res Public Health 10(9):4352–4389
Witham KL, Minchin RF, Butcher NJ (2016) Role for human arylamine N-acetyltransferase 1 in the methionine salvage pathway. Biochem Pharmacol 125:93–100
Komiya Y, Habas R (2008) Wnt signal transduction pathways. Organ 4(2):68–75
Wu G, Huang X, Hua Y, Mu D (2011) Roles of planar cell polarity pathways in the development of neural tube defects. J Biomed Sci 18:66
Claudia Kappen, Anne M. Molloy, Diana M. Juriloff, Muriel J. Harris, (2012) A consideration of the evidence that genetic defects in planar cell polarity contribute to the etiology of human neural tube defects. Birth Defects Research Part A: Clinical and Molecular Teratology 94 (10):824-840
Allache R, De Marco P, Merello E, Capra V, Kibar Z (2012) Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res A Clin Mol Teratol 94(3):176–181
Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene NDE, Copp AJ, Stanier P (2012) Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat 33(2):440–447
Lei Y, Zhu H, Duhon C, Yang W, Ross ME, Shaw GM, Finnell RH (2013) Mutations in planar cell polarity gene SCRIB are associated with spina bifida. PLoS One 8(7):e69262
Wang M, De Marco P, Merello E, Drapeau P, Capra V, Kibar Z (2015) Role of the planar cell polarity gene protein tyrosine kinase 7 in neural tube defects in humans. Birth Defects Res A Clin Mol Teratol. 103(12):1021–1027
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173–1178
Acknowledgments
We would like to acknowledge High Performance Computing Center of University of Tartu. This study was funded by EU H2020 grants 692145, 676550, 654248, the Estonian Research Council Grant IUT20-60 and European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012 GENTRANSMED).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval for the study was obtained from the Ethics Review Committee on Human Research of the University of Tartu. All participants signed an informed consent form prior to enrolment in this study. For minors, consent was obtained from their parents.
Conflict of interest
The authors of this article have no conflict of interest to declare.
Electronic supplementary material
Table S1
(DOCX 35 kb)
Rights and permissions
About this article
Cite this article
Pappa, L., Kals, M., Kivistik, P.A. et al. Exome analysis in an Estonian multiplex family with neural tube defects—a case report. Childs Nerv Syst 33, 1575–1581 (2017). https://doi.org/10.1007/s00381-017-3491-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00381-017-3491-1