Abstract
Sugarcane is an important crop worldwide, and most sugar is derived directly from sugarcane. Due to its thermophilic nature, the yield of sugarcane is largely influenced by extreme climate conditions, especially cold stress. Therefore, the development of sugarcane with improved cold tolerance is an important goal. However, little is known about the multiple mechanisms underlying cold acclimation at the bud stage in sugarcane. In this study, we emphasized that sensitivity to cold stress was higher for the sugarcane variety ROC22 than for GT42, as determined by physical signs, including bud growth capacity, relative conductivity, malonaldehyde contents, and soluble sugar contents. To understand the factors contributing to the difference in cold tolerance between ROC22 and GT42, comparative transcriptome analyses were performed. We found that genes involved in the regulation of the stability of the membrane system were the relative determinants of difference in cold tolerance. Additionally, genes related to protein kinase activity, starch metabolism, and calcium signal transduction were associated with cold tolerance. Finally, 25 candidate genes, including 23 variety-specific and 2 common genes, and 7 transcription factors were screened out for understanding the possible cold resistance mechanism. The findings of this study provide candidate gene resources for cold resistance and will improve our understanding of the regulation of cold tolerance at the bud stage in sugarcane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The frequency of extreme weather, including low temperatures, flooding, waterlogging, and drought (high temperatures), has recently increased. Among these abiotic stresses, cold stress (i.e., chilling and freezing) is particularly common, and the average minimum temperature of approximately 64% of the world total land is under 0 °C (Rihan et al. 2017). Cold stress mainly influences plant growth and development and reduces crop yields and/or the quantity of other useful materials (Pearce 2001). Some plants have evolved mechanisms to withstand cold stress. Under cold stress, physiological, biochemical, and cellular changes take place in plant cells. At the physiological and cellular levels, the osmotic potential, ice crystal formation, cell membrane stability, and concentration and distribution of reactive oxygen species (ROS) are altered substantially (Dong et al. 2009). Moreover, the soluble sugar concentration, cold resistance proteins, and proline content, which are important indexes of cold tolerance in plant cells, clearly change (Kaplan et al. 2007). A reduction in plasma membrane fluidity appears to be a primary event in cold signal transduction (Ding et al. 2019).
In molecular aspect, the well-understood cold-signaling transduction pathway is the CBF-COR transcriptional cascade (Liu et al. 2019). This pathway plays important roles in the promotion of several downstream cold-regulated (COR) genes (Thomashow 2010). Another crucial component of the CBF-COR cascade is the C-repeat binding factors (CBFs) gene family, which known as central nodes in the cold acclimation pathway and is conserved in many plant species. The basic helix-loop-helix transcriptional activator inducer of CBF expression 1 (ICE1) has been identified as a transcriptional activator. It can bind to MYC cis-elements within the CBF3 promoter, thereby repressing the expression of CBF3 and its target COR genes during cold acclimation. In addition, overexpression of ICE1 in Arabidopsis results in increased cold resistance, suggesting that ICE1 has a regulatory role in the cold stress response (Chinnusamy et al. 2003).
Sugarcane is an important crop with high economic and energy-related value (Li et al. 2017). In Southeast Asia, sugarcane is the main sugar crop (Zhang et al. 2011). In China, 60% of sugarcane is produced in Guangxi province. As a tropical crop, sugarcane is sowed in the spring (from late January to mid-march) in most production areas. However, freeze injury during the wintering stage can destroy the growth and development of sugarcane, thereby decreasing sugar production. For example, in 2008, freezing injury over a 23-day period (< 4 °C) was recorded in Guangxi province during the wintering stage, leading to major losses in some sugarcane varieties, especially ROC22. In January of 2018, a 5-day chilling period resulted in a reduction in sugar production in Guangxi province (Huang et al. 2019). During the last few decades, climate change, especially in southern China, the frequency of abnormal conditions is increasing. Chilling and freezing disasters are growing more frequent and unpredictable, resulting in high variation in production and economic losses. Thus, a comprehensive understanding of the mechanism underlying cold tolerance is necessary for the prompt development of solutions.
Several regulators of cold tolerance in sugarcane have been identified. For example, a NAC domain protein family member, SsNAC23, plays roles in the response to cold stress in sugarcane (Nogueira et al., 2005). The overexpression of the sugarcane alcohol dehydrogenase gene family member ScADH3 enhances the cold tolerance of transgenic Nicotiana benthamiana (Su et al. 2020). Furthermore, an uncharacterized gene encoding a cold-regulated protein (Cor413) in Saccharum spontaneum is upregulated in leaf and root tissues under low temperatures (Dharshini et al. 2020). Generally, sugarcane is considered sensitive to low temperatures; however, field observations have shown that the degree of sensitivity varies among varieties (Nogueira et al. 2003a, b). However, these limited information is still insufficient for us to establish a systematic and complete recognition of cold tolerance mechanism in sugarcane. Thus, genetic resources related to cold tolerance can be detected by comparative analyses of different sugarcane varieties.
Although several genes involved in the regulation of cold responses in sugarcane have been identified at different growth stages, few studies have evaluated cold-response genes during the bud stage, despite the important implications in the context of climate change. To address this gap in knowledge, we performed a comparative transcriptome assay using different sugarcane bud materials. Our results provide useful candidate genes for improving the cold resistance of sugarcane at the bud stage.
Materials and Methods
Plant Materials, Treatments, and Sample Collections
Sugarcane varieties ROC22 and GT42 were used as plant materials in this study. ROC22 is the main cultivar in Guangxi province, China. GT42 is introduced from Taiwan, China. The production and saccharinity in GT42 overmatch those in ROC22 (Deng et al. 2018). The culture method was mainly referenced the previous study (Yang et al. 2016; Qiu et al. 2019) with slight modifications. The individual stems of ROC22 and GT42 were cut to be one-bud node and immersed with flowing water for 24 h to promote the sprout of buds and remove impurities, and then cultured in sand soil in the growth chamber (MLR-352H-PC, Panasonic, Japan) at 28 °C with the continuous illumination (30,000 lx) and 65% relative humidity. After the buds grown out and the length reached 4 cm, both of ROC22 and GT42 buds were transported into a low-temperature (4 °C) growth chamber under the continuous illumination (30,000 lx), and the cold treatment maintained 48 h. The control plants were cultured under same condition except the culture temperature was 28 °C. The young buds were collected for the subsequent experiments. For the RNA sequencing sample collections, five buds of control group and cold treated group at 0 h, 24 h, and 48 h were collected using sterilized forceps and knifes, and frozen in liquid nitrogen immediately. Each of RNA-seq sample contained eight buds and was performed three biological replications. For the measurement of physiological indexes, five buds of ROC22 and GT42 at 0 h, 24 h, and 48 h post treatments were collected and stored in − 80 °C refrigerator or used for metrical experiments immediately.
Measurements of Morphological and Physiological Indexes
To monitor the growth status of ROC22 and GT42 under cold treatment, the lengths of buds were measured. The measurements of weights and lengths of buds were performed by measuring 20 buds of ROC22 and GT42 under cold treatment at 0 h, 24 h, and 48 h, respectively. To understand the physiological changes of ROC22 and GT42 under cold treatment, the relative conductivity (a physiological index that reflects cell membrane permeability under stress condition) and malondialdehyde (MDA) content were measured using the relative methods described previously (Li et al. 2008; Ranieri et al. 1993). For relative conductivity measurement, five buds of each time point were cleaned using distilled water, and then dry using absorbing paper. The buds were cut as 1 cm segments and immersed in 20 ml distilled water, and then were vacuumed for 8 min. The conductivity (recorded as S1) was measured using conduct meter at 25 °C. Finally, the bud tissues with distilled water were boiled for 15 min, and then measured the conductivity again (recorded as S2). The relative conductivity was the value of S1/S2. For MDA measurement, five buds of each time point were collected and ground in mortar with liquid nitrogen. Then, 1 g of tissue powder was suspended in 8 ml trichloroacetic acid (10%, v/v), and centrifuged for 10 min under 4000 rpm. The absorbency of supernate was measured using spectrophotometer. The calculation method was referenced as the description in the citing article. Meanwhile, the soluble sugar contents in bud tissues were also measured using anthrone method and the metrical method was described previously (Zou 1995), and five buds of each time point were used. All experiments were replicated three times.
Library Construction and Sequencing
Total RNA was extracted from the bud tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA integrity was also checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). All samples had RNA integrity number (RIN) above 7.0. A total amount of 1.0 μg RNA per sample was used to prepare sequencing libraries using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). An Illumina HiSeq 2000 platform was used for sequencing, and 150 bp paired-end reads were generated.
The Quality Control of Transcriptomic Data and Mapping on the Reference Genome
The total raw sequencing data were filtered by Phred quality score (Q ≥ 30) and read length (≥ 25 bp) using the SolexaQA package. The reads containing adapters and poly-N, and low-quality reads were removed from the raw reads. The clean reads were mapped to the reference sugarcane genome (http://sugarcane.zhangjisenlab.cn/sgd/download/Sspon.HiC_chr_asm.fasta.gz) (Zhang et al. 2018) using TopHat2 (Kim et al. 2013). The sugarcane genome assembly and gene annotation are deposited in the NCBI database under accession number QVOL00000000, BioProject number PRJNA483885 and BioSample number SAMN09753102 (Zhang et al. 2018).
Isolation and Functional Analyses of DEGs
The expression level of each gene were quantified based on the mapped reads counts using featureCounts software (v 2.0.1) with default parameters. For biologically duplicated samples, we performed an analysis using DESeq2 to screen differential expression genes (DEGs) (Anders and Huber 2010). The screening threshold were expression fold change ≥ 1 and a false discovery rate (FDR) < 0.01. The published sugarcane gene annotation was referenced to annotate all DEGs (Zhang et al. 2018). Gene Ontology (GO) enrichment analysis was performed using topGO (v 2.42.0) (corrected p-value < 0.05) (Alexa and Rahnenfuhrer 2020). The DEGs that were gained from control groups of ROC22 and GT42 were thought as developmental background and removed from DEG sets isolated from cold treated groups.
Gene Validation Using Quantitative Real-Time PCR
The First-strand cDNA synthesis was performed using M-MLV Reverse Transcriptase (Takara, Japan) according the protocol. The CFX96 Touch™ Real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) was used for DEGs expression analysis using the SYBR Premix Ex Taq Kit (Takara, Japan). The thermal cycling program was used for PCR: 45 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s after an initial denaturation step for 5 min at 95 °C. The 2−ΔΔCT method was used to calculate gene expression. Each experiments performed three biological replications. The sugarcane glyceraldehyde-phosphate dehydrogenase (GAPDH) gene was used as the inner reference (Thiebaut et al. 2012), and all primers used in this study were listed in Table S1.
Statistical Calculation and Data Availability
SPSS 18.0 software was used to perform statistical calculation. One-way ANOVA with post hoc Tukey HSD (Honestly Significant Difference) test (p < 0.05) was carried out to detect the differences among different data sets. The RNA sequencing data sets were uploaded in Sequence Read Archive (SRA) database with the BioProject Accession Number PPJNA667204 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA667204).
Results
ROC22 and GT42 Show Different Cold Sensitivity at the Bud Stage
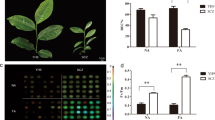
Previous studies have shown that the sugarcane variety ROC22 is highly sensitive to low temperatures (Sun et al. 2012); however, little is known about this trait in GT42. To better understand the differences between ROC22 and GT42 under low-temperature treatment during the bud stage, we monitored the bud growth status at different time points. Firstly, we observed the morphological changes of buds of both ROC22 and GT42. Under normal condition, the growth of ROC22 and GT42 buds grew normally, and both of them started leaves at 48 h. After cold treatment, the growth of ROC22 was inhibited, while the growth of GT42 buds were similar as buds grown under normal condition (Fig. 1A). To quantify these growth differences, we next measured the fresh weights and lengths of buds. Comparing with the buds grown under normal condition, the fresh weights and lengths of ROS22 and GT42 buds were both decreased, indicating that the growth of ROC22 and GT42 was restrained under cold treatment (Fig. 1B, C). However, GT42 displayed higher relative growth rate due to GT42 showed obviously higher fresh weight and longer length after cold treatment (Fig. 1B, C).
Changes of growth and physiological indexes of ROC22 and GT42 buds. A Morphological observations of ROC22 and GT42 buds under normal and cold conditions. Scale bars: 1 cm; B fresh weights of ROC22 and GT42 buds under normal and cold conditions. C Bud lengths of ROC22 and GT42 under normal and cold conditions; D relative conductivity of ROC22 and GT42 buds under normal and cold conditions; E MDA contents of ROC22 and GT42 buds under normal and cold conditions. F Soluble sugar contents in ROC22 and GT42 buds under normal and cold conditions. Data was based on 20 individual samples and One-way ANOVA with post hoc Tukey HSD (Honestly Significant Difference) test (**p < 0.05) was carried out to detect the differences among different data sets. The alphabets indicate differences among different data sets
Plant cell membrane permeability is an important determinant of plant stress resistance, including cold resistance (Liu et al. 2013). To evaluate the physiological changes of cell membrane in ROC22 and GT42 under low-temperature conditions, we evaluated relative conductivities and malonaldehyde (MDA) contents in bud samples at different time points. As shown in Fig. 1D, both ROC22 and GT42 displayed increases in relative conductivity after cold treatment, and ROC22 showed significantly higher relative conductivities than those in GT42. Under normal condition, both ROC22 and GT42 revealed low relative conductivity, and they displayed the similar increased rate of relative conductivity (Fig. 1D). Moreover, we found higher levels of MDA in ROC22 than in GT42 at each time point under cold treatment, while there were no clear differences between different time points under normal condition. (Fig. 1E). As a crucial cellular component and metabolite, soluble sugar also play key roles in plants to against cold stress. Thus, we next measured the soluble sugar contents in ROC22 and GT42 buds at each time point under normal and cold treatments. The soluble sugar contents in ROC22 and GT42 obviously increased after cold treatment but lower than that at each time point under normal condition. However, the increase in soluble sugar in GT42 was significantly higher than that in ROC22 at 24 and 48 h after cold treatment (Fig. 1F). Taken together, the results shown above indicated that ROC22 and GT42 maintained clear differences in the response to cold stress, and cold resistance was higher in GT42 than in ROC22 at the bud stage.
Summary of Transcriptome Data
The responses of ROC22 and GT42 to cold treatment differed significantly at 24 and 48 h, prompting us to evaluate the genetic factors underlying the response to cold treatment. To clarify the gene expression differences between ROC22 and GT42, we collected bud tissues at 0 h, 24 h, and 48 h after cold treatment and performed transcriptome sequencing. After filtering out adaptors and low-quality reads, transcriptome sequencing of 18 samples generated, on average, 22,291,792 (ROC22, 0 h), 22,359,858 (ROC22, 24 h), 19,943,231 (ROC22, 48 h), 22,584,049 (GT42, 0 h), 22,869,800 (GT42, 24 h), and 23,668,153 (GT42, 48 h) clean reads (Table S2). At least 94.62% of the clean reads showed eligible Q30 (%) values in all libraries, and the GC ratio of clean reads was higher than 54% (Table S2), indicating that our sequencing data were of sufficiently high quality for subsequent analyses. On average, 73.05% of clean reads were mapped to the sugarcane reference genome, and over 94% of mapped clean reads were uniquely mapped (Table S3).
Functional Analyses of Variety-Specific Differentially Expressed Genes (DEGs)
To investigate the effects of cold stress on ROC22 and GT42 from a genetic perspective, we isolated differentially expressed genes (DEGs) at 0 h, 24 h, and 48 h. In total, 703 DEGs, including 44 up- and 659 downregulated genes, were screened out in the 24 h vs. 0 h comparison in the cold-sensitive sugarcane variety ROC22 (Fig. 2A), while 887 DEGs, including 318 up- and 569 downregulated genes, were identified in the 48 h vs. 24 h comparison (Fig. 2A). In the cold-tolerant sugarcane GT42, 53 (35 up- and 19 downregulated) and 356 (10 up- and 346 downregulated) DEGs were identified in the 24 h vs. 0 h and 48 h vs. 24 h comparisons, respectively (Fig. 2B). Since continuous changes in physical signs were observed, we expected DEGs identified in both the 24 h vs. 0 h and 48 h vs. 24 h comparisons to play particularly important roles in the regulation of cold tolerance. Hence, we further isolated 66 and 13 DEGs in the intersection between the 24 h vs. 0 h and 48 h vs. 24 h comparisons in ROC22 and GT42 (Fig. 2C and D). We found that 66 DEGs displayed three different expression patterns in ROC22 during cold treatment (Fig. 2E), while 13 DEGs in GT42 showed two different expression profiles (Fig. 2F).
Comparative RNA-seq analysis of transcriptomes of ROC22 and GT42. A, B Numbers of DEGs in ROC22 and GT42 at each comparable group. The up- and downregulated DEGs were showed; C, D Venn diagrams of common DEGs in ROC22 and GT42, respectively; E, F Heatmap showed the expression patterns of common DEGs in ROC22 and GT42, respectively
To understand the functions of these variety-specific DEGs, a GO enrichment analysis was carried out to filter the genes involved in cold stress-related processes. We found that ROC22-specific DEGs were enriched for the terms starch catabolic process (4 DEGs) and membrane system-related terms, including Golgi membrane (1 DEG), membrane (1 DEG), plasma membrane (3 DEGs), vacuolar membrane (3 DEGs), cytoplasmic membrane-bounded vesicle (4 DEGs), and integral component of membrane (15 DEGs). In addition, 4 and 4 DGEs were related to calcium ion binding and beta-amylase activity, respectively (Fig. 3A). We also found that several GT42-specific DEGs were related to membrane system-related terms, including Golgi membrane (1 DEG), integral component of plasma membrane (1 DEG), cytoplasmic membrane-bounded vesicle (2 DEGs), and integral component of membrane (3 DEGs). Additionally, some GT42-specific DEGs were also involved in the stress-activated protein kinase signaling cascade (1 DEG) and activation of protein kinase activity (1 DEG). One DEG was associated with calcium ion transmembrane transport and one DEG was associated with calcium-transporting ATPase activity (Fig. 3B).
Functional Analyses of Common DEGs in Both ROC22 and GT42
We further evaluated common DEGs with predicted roles in the regulation of cold tolerance in both ROC22 and GT42. We compared variety-special DEGs and screened out six common DEGs (Fig. 4A). Each of these six common DEGs participated in several biological, cellular, and molecular processes. Notably, we found that Sspon.004C0011332 was related to the term ‘Golgi membrane’ and Sspon.004D0012690 was related to the terms ‘integral component of membrane and cytoplasmic membrane-bounded vesicle’ (Fig. 4B, Table 1). These results indicated that cell membrane system-related genes are collectively involved in the regulation of cold resistance in ROC22 and GT42, and the stability of the membrane system might explain the difference in cold resistance between the varieties.
Expression Profiles of Candidate Genes
In addition to broad analyses of similarities and differences in gene modules involved in the regulation of cold tolerance in ROC22 and GT42, detailed analyses of the spatiotemporal expression patterns of candidate genes can provide insight into gene function. Based on functional analyses, we identified 25 candidate genes, including 18 ROC22-specific, 5 GT42-specific, and 2 common DEGs (Table 1). To further investigate the expression patterns of these genes in ROC22 and GT42, we performed qRT-PCR assays. All 18 ROC22-specific genes were upregulated in ROC22, consistent with expression trends determined by RNA-seq data. However, these genes did not display significant changes in expression in GT42 after cold treatment (Fig. 5). Inversely, five GT42-specific genes were upregulated under cold stress but were unchanged in ROC22 (Fig. 5). The two common genes were upregulated; however, levels of both genes were clearly higher in GT42 than in ROC22 (Fig. 5). These results demonstrated that these genes might be involved in cold tolerance in sugarcane via different expression patterns in ROC22 and GT42, and two common genes encoding a B-box zinc finger protein (Sspon.004C0011332) and CMP-sialic acid transporter (Sspon.004D0012690) might be important candidate markers for cold resistance in sugarcane.
Expression patterns of candidate DEGs. qRT-PCR assay was performed to detect the relative expression levels of each candidate DEG. The expression level at 0 h in ROC22 was set as 1. Each of experiment was replicated three times. Bars shown means ± SD. Candidate DEGs in ROC22 were marked by black color. Candidate DEGs in GT42 were marked by red color. Candidate DEGs in both ROC22 and GT42 were denoted by blue color
Isolation of Transcription Factors that Responded to Cold Stress
Transcription factors (TFs) have important functions in the regulation of target genes at the transcriptional level. Several kinds of TFs, such as WRKY, NAC, and ethylene response factors (ERF), have been reported to play essential roles in the regulation of cold resistance. We next surveyed the number and type of TFs among candidate genes. Among ROC22-specific DEGs, five TFs, including two WRKY TFs (Sspon.003C0035890 and Sspon.007A0013201), two NAC TFs (Sspon.002B0025720 and Sspon.003B0009181), and one ERF (Sspon.004C0008870), were screened out (Table 2). Two TFs were identified from GT42-specific DEGs, including one ERF (Sspon.005C0001440) and one C2H2 zinc finger TF (Sspon.001A0036160) (Table 2). There were no TFs in the common DEG set. As determined by qRT-PCR analyses, all five ROC22-specific TFs were upregulated in ROC22 during cold treatment but were not affected in GT42 (Fig. 6). In contrast, GT42-specific ERF and C2H2 TFs were upregulated in GT42 under cold stress and showed no expressed differences in ROC22 (Fig. 6).
Validation of DEGs
To confirm the reliability of the RNA-seq results, 20 randomly selected DEGs in the 24 h vs. 0 h and 48 h vs. 24 h comparisons for ROC22 and GT42 were evaluated by qRT-PCR. As shown in Table 3, all DEGs showed consistent trends, apart from some differences in fold change values. These results indicated that our RNA-seq data were highly reliable.
Discussion
Despite an increasing global temperature, abnormal weather and cold extremes are still observed worldwide (Gupta and Deswal 2014; Horton et al. 2015). Especially in southern China, the frequency of low temperatures is increasing. This is a challenge from the perspective of tropical agriculture. To expand the growth region to high latitudes and altitudes, it is necessary to developed improved cold-tolerant crop varieties (Ding et al. 2019), including important economic and energy-related crops, such as sugarcane. However, our understanding of cold tolerance in sugarcane is still limited, especially at the bud stage. Therefore, uncovering the mechanism underlying cold tolerance and the identification of crucial gene resources are necessary for improving cold acclimation in sugarcane.
Cell Membrane Stability is Essential for Cold Tolerance in Sugarcane Buds
When plants are exposed to cold stress, the cell membrane is thought to be the primary site of injury due to its central role in the regulation of various cellular processes (Takahashi et al. 2013). Therefore, cell membrane stability is a key trait for analyses of abiotic stresses and the selection of tolerant genotypes in plants (ElBasyoni et al. 2017). Under stresses, membrane lipid peroxidation often occurs in the cell membrane, and MDA is a representative product of membrane lipid peroxidation. MDA contents can reflect the extent of injury in the cell membrane (Robert and Bewlery 1980). Membrane lipid peroxidation can also alter membrane conductivity. Under cold conditions, membrane conductivity is often increased in sugarcane (Zhu et al. 2018). In this study, we found that ROC22 was more sensitive to cold stimulus than GT42, as evidenced by the better growth of GT42 buds under cold stress (Fig. 1A). In addition, GT42 showed lower increases in relative conductivity and MDA contents than those of ROC22 (Fig. 1B, C). A previous study has reported that cell membrane stability is greater in plant varieties with high cold resistance (Zhu et al. 2018). Our results are consistent with this finding, indicating that cell membrane stability is essential for cold tolerance in sugarcane buds.
We also found similarities in the molecular mechanisms underlying cold resistance in ROC22 and GT42. Several DEGs, including phylloplanin, AAA-ATPase ASD, and sodium/hydrogen exchanger 2 encoding genes, were involved in processes related to the cell membrane in both ROC22 and GT42 (Figs. 3, 4B, Table 1). Furthermore, we found that the calcium-related GO terms calcium ion binding, calcium ion transmembrane transport, and calcium-transporting ATPase activity are important in ROC22 and GT42 (Fig. 3), indicating that the calcium ion transmembrane transport capacity and calcium-dependent processes are altered under cold stress. These results provide insights at the genetic level and suggest that cell membrane stability is the core determinant of differences of cold resistance between ROC22 and GT42. Two common genes encoding a B-box zinc finger protein 24 (Sspon.004C0011332) and a CMP-sialic acid transporter 5 (Sspon.004D0012690) may play key roles in the regulation of cell membrane stability in sugarcane. The possible functions of these relative DEGs are in line with our observations of physical signs.
We also observed differences between the strains. Several variety-specific DEGs contributed to the response to cold stress. For example, we isolated a gene encoding mitogen-activated protein kinase kinase A (MAPKK; Sspon.003C0017630) in GT42 but not in ROC22 (Fig. 3, Table 1). Its expression levels are increased under cold stress in GT42 but are not affected by cold in ROC22 (Fig. 5). Given the essential roles of the MAPK cascade in several stresses (Chen et al. 2021), MAPK-mediated pathways may contribute to the increase in cold resistance in GT42, and these pathways are highly correlated with cell membrane stability. Taken together, our data reveal that cell membrane stability is an important indicator of cold resistance, and the candidate genes can be used to improving cold tolerance in sugarcane at the seeding stage.
Soluble Sugar and Starch Metabolism May Influence Cold Tolerance in Sugarcane Buds
Soluble sugars exert protective effects on plant cells against damage caused by cold stress by many mechanisms (e.g., by acting as osmoprotectants or nutrients as well as by interacting with the lipid bilayer) (Ma et al. 2009). Several soluble sugars, such as sucrose, glucose, fructose, ribose, and trehalose, often accumulate under low-temperature stress (Wei et al. 2019). These kinds of sugars function in free radical clearance and indirectly induce protein synthesis, thereby altering the cold tolerance of plants (Selvarajan et al. 2018). In our study, we found that the soluble sugar contents in ROC22 and GT42 differed significantly under cold treatment (Fig. 1D). Consistent with this result, we found that several DEGs were enriched in beta-amylase activity in ROC22 (Fig. 3, Table 1), indicating that starch metabolism is a variety-specific process involved in regulating cold tolerance in sugarcane. As a storage polysaccharide, starch is found in sugarcane stalks, leaves and growing point regions (de Figueira et al. 2011). The accumulation of soluble sugars may be at least partially triggered by an increased rate of starch degradation under cold treatment (Sitnicka and Orzechowski 2014); thereby, starch metabolism is thought to be closely related to tolerance against multiple stresses (Krasensky and Jonak 2012). Four genes encoding beta-amylase 3 were isolated, and Sspon.001A0018210 and Sspon.001B0023140 showed clear differences in expression levels under cold stress (Fig. 5), indicating that these two genes may be possible factors involved in regulating starch metabolism.
Transcription Factors May Play Roles in Regulating Cold Resistance in Sugarcane Buds
TFs are regulated by different signal transduction pathways and can directly or indirectly bind to cis-acting elements to alter the transcription efficiency of target genes related to plant growth, development, and stress responses. In this study, we found that some WRKY TFs were involved in the response to cold stress in ROC22 (Table 2). These two putative WRKY genes (Sspon.003C0035890 and Sspon.007A0013201) were upregulated in ROC22 but did not change in GT42 after cold treatment (Fig. 6). WRKY TFs, which are among the well-characterized class of TFs in plants, have reported roles in cold tolerance, and 39 WRKY TFs have been isolated in sugarcane (Javed et al. 2020). Proteins in the NAC TF family are specific to plants and play important roles in plant growth, development, and senescence (Kikuchi et al. 2000). In a pioneering study, three putative NAC TFs were identified as cold-response genes in sugarcane (Nogueira et al. 2003a, b). Another NAC TF, SsNAC23, is induced by low temperatures in sugarcane (Ditt et al. 2011). Here, we found that the expression levels of two NAC TFs increased by cold treatment in ROC22 but remained unchanged in GT42 (Table 2, Fig. 6). ERFs are a plant-specific TF family that was initially identified in tobacco as binding proteins for reduced sensitivity to disease (Wang et al. 2014). Increasing studies have revealed that ERFs play key roles in plant stress responses (Wang et al. 2014). SodERF3, a sugarcane ERF, enhances drought and osmotic stress tolerance in transgenic tobacco (Trujillo et al. 2008). However, the functions of ERFs in cold resistance in sugarcane are largely unknown. In this study, we isolated one ERF from ROC22 and one ERF from GT42 (Table 2) and observed distinct expression profiles in the two varieties (Fig. 6), suggesting that these two ERFs function in a variety-specific manner. More important, cold stress may result in water shortage, therefor give rise to the expression differences of these ERFs. Meanwhile, we also screened out a C2H2 zinc finger TF from GT42 (Table 2) and found that it was a cold-response protein in GT42 but insensitive to cold in ROC22 (Fig. 6). So far, there are very limited information of the biological roles of C2H2 in regulating cold tolerance in sugarcane. Thus, our data provide novel insights into the molecular mechanisms underlying cold tolerance in sugarcane during bud stage.
Conclusion
In this study, we confirmed that the sugarcane variety ROC22 is more sensitive than GT42 by monitoring physical signs under cold treatment during the bud stage. Under cold stress, GT42 buds show higher fresh weights and lengths. The results of relative conductivity and MDA contents reveal that GT42 buds have stronger stabilities of cell membrane under cold treatment. Through performing a comparative transcriptome analysis using the bud tissues, we identified candidate genes involved in the regulation of cold acclimation at the bud stage. The candidate genes, which involved in cell membrane stability, soluble sugar metabolism, and starch metabolism, support us new insights into cell membrane-based regulatory mechanism of cold tolerance in sugarcane. Two common genes, which encode B-box zinc finger protein 24 and CMP-sialic acid transporter 5, may be potential candidates involved in cold resistance regulation. More importantly, TFs isolated in this study, including WRKY, NAC, ERF, and C2H2, may regulate these two gene clusters at the transcriptional level. Interactions among these candidate genes may have synergistic effects on cold resistance during sugarcane bud stage, and the possible functions of these candidates should be studied from genetic aspect in future. Based on these findings, we speculate that the TFs including WRKY, NAC, ERF, and C2H2 may synchronously regulate the cell membrane stability and soluble sugar and starch metabolisms to response cold stress, in turn to control cold resistance in sugarcane from transcriptional control aspect during bud stage, and more verified works need to be done in the future (Fig. 7). Overall, our results provide useful information on differentially expressed gene modules and metabolic and signaling pathways related to the response to cold stress in sugarcane during bud stage. Our data also provide a genetic resource for studies of cold tolerance regulatory genes and for sugarcane breeding.
Mechanism adapted to combat low-temperature stress conditions. Based on the results in this study, the cold tolerance in sugarcane at bud stage established on a complex network which several genes involved in. In this putative network, genes that involved in cell membrane system and soluble sugar and starch metabolism were isolated and may play roles in regulating cold resistance in sugarcane buds. Moreover, transcription factors may also played important roles in regulating these two gene clusters synergistically. Solid line arrows represent the direct regulation and dashed-line arrows represent the putative regulation
References
Alexa A, Rahnenfuhrer J (2020). topGO: enrichment analysis for gene ontology. R package version 2.42.0
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Chen X, Ding Y, Yang Y, Song C, Wang B, Yang S, Guo Y, Gong Z (2021) Protein kinases in plant responses to drought, salt, and cold stress. J Integr Plant Biol 63:53–78
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
de Figueira JA, Carvalho PH, Sato HH (2011) Sugarcane starch: quantitative determination and characterization. Food Sci Technol (campinas) 31:806–815
Deng Y, Wang L, Wang Z, Huang H, Xian W (2018) Analysis and evaluation for varietal characteristics and high yield stability of sugarcane variety Guitang 42. J South Agric 49:42–47
Dharshini S, Manoj VM, Suresha GS, Narayan JA, Padmanabhan TSS, Kumar R, Meena MR, Manickavasagam M, Ram B, Appunu C (2020) Isolation and characterization of nuclear localized abiotic stress responsive cold regulated gene 413 (SsCor413) from Saccharum spontaneum. Plant Mol Biol Rep 38:628–640
Ding YL, Shi YT, Yang SH (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222:1690–1704
Ditt RF, Gentile A, Tavares RG, Camargo SR, Fernandez JH, Da Silva MJ et al (2011) Analysis of the stress-inducible transcription factor SsNAC23 in sugarcane plants. Sci Agric 68:454–461
Dong CH, Zolman BK, Bartel B, Lee BH, Stevenson B, Agarwal M, Zhu JK (2009) Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol Plant 2:59–72
ElBasyoni I, Saadalla M, Baenziger S, Bochelman H, Sabah M (2017) Cell membrane stability and association mapping for drought and heat tolerance in a worldwide wheat collection. Sustainability 9:1606
Gupta R, Deswal R (2014) Antifreeze proteins enable plants to survive in freezing conditions. J Biosci 39:931–944
Horton DE, Johnson NC, Singh D, Swain DL, Rajaratnam B, Diffenbaugh NS (2015) Contribution of changes in atmospheric circulation patterns to extreme temperature trends. Nature 522:465–469
Huang YH, Kuang ZM, Li L, Tan MX, Ou ZR, Liu ZP (2019) Quantitative assessment of the impact of low temperature, rain and snow freezing weather on sugarcane in Guangxi in January 2018. J Meteorol Res Appl 40:65–67
Javed T, Shabbir R, Ali A, Afzal I, Zaheer U, Gao SJ (2020) Transcription factors in plant stress responses: challenges and potential for sugarcane improvement. Plants 9:491
Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50:967–981
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano H-Y (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 262:1047–1051
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608
Li GW, Zhang MH, Cai WM, Sun WN, Su WA (2008) Characterization of OsPIP2;7, a water channel protein in rice. Plant Cell Physiol 49:1851–1858
Li M, Liang Z, He SS, Zeng Y, Jing Y, Fang WK, Wu KC, Wang GY, Ning X, Wang LW, Li S, Tan HW, Tan F (2017) Genome-wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.). BMC Genomics 18:754
Liu W, Yu K, He T, Li F, Zhang D, Liu J (2013) The low temperature induced physiological responses of Avena nuda L., a cold-tolerant plant species. Sci World J 2013:658793
Liu Y, Dang P, Liu L, He C (2019) Cold acclimation by the CBF–COR pathway in a changing climate: lessons from Arabidopsis thaliana. Plant Cell Rep 38:511–519
Ma YY, Zhang YL, Lu J, Shao HB (2009) Roles of plant soluble sugars and their responses to plant cold stress. Afr J Biotechnol 8:2004–2010
Nogueira FTS, De Rosa JVE, Menossi M, Ulian EC, Arruda P (2003a) RNA expression profiles and data mining of sugarcane response to low temperature. Plant Physiol 132:1811–1824
Nogueira FTS, De Rosa VE, Menossi M, Ulian EC, Arruda P (2003b) RNA expression profiles and data mining of sugarcane response to cold temperature. Plant Physiol 132:1811–1824
Nogueira FTS, Schlögl PS, Camargo SR, Fernandez JH, De Rosa JVE, Pompermayer P, Arruda P (2005) SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Sci 169:93–106
Pearce RS (2001) Plant freezing and damage. Ann Bot 87:417–424
Qiu LH, Chen RF, Fan YG, Huang X, Luo HM, Xiong FQ, Liu JX, Zhang RH, Lei JC, Zhou HW, Wu JM, Li YR (2019) Integrated mRNA and small RNA sequencing reveals microRNA regulatory network associated with internode elongation in sugarcane (Saccharum officinarum L.). BMC Genomics 20:817
Ranieri A, Lencioni L, Schenone G, Soldatini GF (1993) Glutathione-ascorbic acid cycle in pumpkin plants grown under polluted air in open-top chambers. J Plant Physiol 142:286–290
Rihan HZ, Al-Issawi M, Fuller MP (2017) Advances in physiological and molecular aspects of plant cold tolerance. J Plant Interact 12:143–157
Robert RC, Bewlery JD (1980) Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol 65:245–248
Selvarajan D, Mohan CM, Dhandapani V, Nerkar G, Jayanarayanan AN, Mohanan MV, Murugan N, Kaur L, Chennappa M, Kumar R, Meena M, Ram B, Chinnaswamy A (2018) Differential gene expression profiling through transcriptome approach of Saccharum spontaneum L. under low temperature stress reveals genes potentially involved in cold acclimation. 3 Biotech 8:195
Sitnicka D, Orzechowski S (2014) Cold-induced starch degradation in potato leaves—intercultivar differences in the gene expression and activity of key enzymes. Biol Plant 58:659–666
Su WH, Ren YJ, Wang DJ, Su YC, Feng JF, Zhang C, Tang HC, Xu LP, Muhammad KS, Que YX (2020) The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation. BMC Genomics 21:521
Sun F, Yang LT, Xie XN, Liu GL, Li YR (2012) Effect of chilling stress on physiological metabolism in chloroplasts of seedlings of sugarcane varieties with different chilling resistance. Acta Agron Sin 38:732–739
Takahashi D, Li B, Nakayama T, Kawamura Y, Uemura M (2013) Plant plasma membrane proteomics for improving cold tolerance. Front Plant Sci 4:90
Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CRC, de Engler JA, Hemerly AS, Ferreira PCG (2012) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35:502–512
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577
Trujillo LE, Sotolongo M, Menendez C, Ochogava ME, Coll Y, Hernandez I et al (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49:512–515
Wang S, Yao W, Wei H, Jiang T, Zhou B (2014) Expression patterns of ERF genes underlying abiotic stresses in di-haploid Populus simonii × P. nigra. Sci World J 2014:745091
Wei TW, Wang Y, Xie ZZ, Guo DY, Chen CW, Fan QJ, Deng XD, Liu JH (2019) Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata. Plant Biotechnol J 17:1394–1407
Yang Y, Zhang X, Chen Y, Guo J, Ling H, Gao S, Su Y, Que Y, Xu L (2016) Selection of reference genes for normalization of MicroRNA expression by RT-qPCR in sugarcane buds under cold stress. Front Plant Sci 7:86
Zhang BQ, Yang LT, Li YR (2011) Comparison of physiological and biochemical characteristics related to cold resistance in sugarcane under filed conditions. Acta Agron Sin 37:496–505
Zhang J, Zhang X, Tang H, Zhang Q et al (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50:1565–1573
Zhu P, Pang X, Liang C, Tan Q, Yan L, Zhou Q, Ou K (2018) Effects of cold stress on reactive oxygen metabolism and antioxidant enzyme activities of sugarcane seedlings. Crops 4:131–137
Zou Q (1995) Plant physiological and biochemical experiment guidance. China Agricultural Press, Beijing
Acknowledgements
We would like to acknowledge the financial support provided by the National Natural Science Foundation of China (31660356), the Development and Regulation of Economically Important Traits in Tropical Crops (2018YFD1000500), the Project funded by Guangxi Postdoctoral Science Foundation, Guangxi Science and Technology Base and Talents Special Project (GuikeAD17195100), the Fund for Guangxi Innovation Teams of Modern Agriculture Technology (nycytxgxcxtd-03-01), the Natural Science Foundation of Guangxi Province (2015GXNSFBA139011), and the Fund for Science and Technology Development of Guangxi Academy of Agricultural Sciences (Guinongke2017JZ19, Guinongke2016JM06, Guinongke2021YT013).
Author information
Authors and Affiliations
Contributions
XH and JW designed the experiment. XH, BZ, XS, YL, CL, ZQ and DL performed the fieldwork and laboratory experiments together. XH and YL wrote the main part of the manuscript together. YL and JW revised the manuscript. All authors reviewed the manuscript, added details to it, and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Handling Editor: Saddam Hussain.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, X., Liang, Y., Zhang, B. et al. Comparative Transcriptome Analysis Reveals Potential Gene Modules Associated with Cold Tolerance in Sugarcane (Saccharum officinarum L.). J Plant Growth Regul 41, 2614–2628 (2022). https://doi.org/10.1007/s00344-021-10437-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10437-9