Abstract
Objective
To investigate the effect of uncertainty estimation on the performance of a Deep Learning (DL) algorithm for estimating malignancy risk of pulmonary nodules.
Methods and materials
In this retrospective study, we integrated an uncertainty estimation method into a previously developed DL algorithm for nodule malignancy risk estimation. Uncertainty thresholds were developed using CT data from the Danish Lung Cancer Screening Trial (DLCST), containing 883 nodules (65 malignant) collected between 2004 and 2010. We used thresholds on the 90th and 95th percentiles of the uncertainty score distribution to categorize nodules into certain and uncertain groups. External validation was performed on clinical CT data from a tertiary academic center containing 374 nodules (207 malignant) collected between 2004 and 2012. DL performance was measured using area under the ROC curve (AUC) for the full set of nodules, for the certain cases and for the uncertain cases. Additionally, nodule characteristics were compared to identify trends for inducing uncertainty.
Results
The DL algorithm performed significantly worse in the uncertain group compared to the certain group of DLCST (AUC 0.62 (95% CI: 0.49, 0.76) vs 0.93 (95% CI: 0.88, 0.97); p < .001) and the clinical dataset (AUC 0.62 (95% CI: 0.50, 0.73) vs 0.90 (95% CI: 0.86, 0.94); p < .001). The uncertain group included larger benign nodules as well as more part-solid and non-solid nodules than the certain group.
Conclusion
The integrated uncertainty estimation showed excellent performance for identifying uncertain cases in which the DL-based nodule malignancy risk estimation algorithm had significantly worse performance.
Clinical relevance statement
Deep Learning algorithms often lack the ability to gauge and communicate uncertainty. For safe clinical implementation, uncertainty estimation is of pivotal importance to identify cases where the deep learning algorithm harbors doubt in its prediction.
Key Points
• Deep learning (DL) algorithms often lack uncertainty estimation, which potentially reduce the risk of errors and improve safety during clinical adoption of the DL algorithm.
• Uncertainty estimation identifies pulmonary nodules in which the discriminative performance of the DL algorithm is significantly worse.
• Uncertainty estimation can further enhance the benefits of the DL algorithm and improve its safety and trustworthiness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The National Lung Screening Trial (NLST) and Dutch-Belgian NELSON lung cancer screening trial provide evidence that lung cancer mortality can be reduced by repeated screening of high-risk individuals with low-dose chest CT [1, 2]. This decrease is primarily attributed to early detection of lung cancer in stages I and II, where a favorable prognosis is more probable than in stages III or IV [3]. Lung cancer screening could therefore play a crucial role in reducing lung cancer mortality. However, a global shortage of radiologists can still lead to a delayed diagnosis [4].
Deep learning (DL) can be a helpful tool to contribute to reducing radiologists’ workload and assist radiologists in lung cancer diagnosis. Venkadesh et al proposed a DL algorithm for pulmonary nodule malignancy risk estimation that was internally and externally validated on data from the NLST and Danish Lung Cancer Screening Trial (DLCST), respectively [5, 6]. The algorithm showed excellent performance on par with thoracic radiologists for pulmonary nodule malignancy risk estimation in an external validation set [6].
An imperative aspect of the safe clinical implementation of such DL algorithms is the ability to gauge and communicate uncertainty [7]. Estimating uncertainty is crucial to identify situations where the algorithm harbors doubt about its predictions and helps identify cases prone to potential DL interpretation errors [8]. Leveraging this uncertainty estimation could optimize the clinical workflow by preventing the algorithm from making a diagnosis in cases of high uncertainty and referring these to clinical experts for further evaluation. Conversely, clinical experts may have more confidence in the algorithm’s prediction when certainty is high. This proactive integration of uncertainty estimates holds promise to minimize error risks, thereby elevating the safety profile of a clinically adopted algorithm [9]. Existing studies corroborate the efficacy of uncertainty estimation in filtering out ambiguous cases, thereby enhancing algorithm accuracy in the remaining cases [10,11,12,13,14,15,16]. However, while most studies primarily focus on refining algorithm accuracy and not on evaluating causes of uncertainty, a prevailing limitation lies in the inadequate identification of underlying trends responsible for inducing uncertainty. Addressing this limitation is important because without understanding what causes uncertainty, it is challenging to improve the reliability of a DL algorithm.
In this study, we build upon a previously developed DL algorithm and integrate a method for uncertainty estimation. Our aim was to investigate the performance of the algorithm for pulmonary nodule malignancy risk estimation when different uncertainty thresholds are applied and identify nodule characteristics responsible for inducing uncertainty in the algorithm.
Methods
Data collection
This retrospective study was conducted using data from two data sources: the Danish lung cancer screening trial (DLCST) [5] and a Dutch tertiary academic center (clinical dataset). DLCST was used as a development set for the uncertainty estimation method. The DLCST dataset consists of anonymized low-dose chest CT examinations that were acquired from participants in the screening trial between 2004 and 2010. Approval for DLCST was obtained from the Ethics Committee of Copenhagen County, and all participants provided informed consent. Two thoracic radiologists performed all nodule annotations in DLCST. The malignant status of nodules was confirmed by histological analysis, and for benign status, stability over at least 2 years of CT follow-up was used. For participants diagnosed with lung cancer, we included the first image on which the malignant nodule was annotated. For participants without a lung cancer diagnosis, we included nodule annotations from the baseline image.
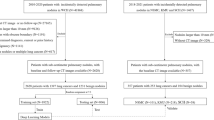
The clinical dataset served as an external validation set, which consists of chest CT examinations in individuals aged 40 years and older acquired for clinical indications, with diverse CT scan protocols (with/without iodine contrast) between 2004 and 2012. The institutional review board waived the need for informed consent because of the retrospective design and data pseudonymization. Subjects were linked to the Dutch national cancer registry to gather information on lung cancer diagnosis until the end of 2014. Further details of the collection of the dataset are described in Chung et al (Thorax, 2018) [17]. We adjusted the set of malignant and benign nodules from Chung et al. by excluding all nodules from patients with a history of other primary cancers. We added this exclusion criterion to remove any participants with any potential metastases. For the clinical dataset, the malignant status of nodules was also confirmed by histological analysis, and for benign status, stability with a median of 5+ years of CT follow-up was used. A flow chart of the data collection and selection is shown in Figure 1 for both datasets.
Flow chart of the data collection and selection of pulmonary nodules. a Nodules from the DLCST were used for the development of the uncertainty estimations. b Incidental nodules from the clinical dataset for validation of the uncertainty estimations. Retrieval errors may be due to anonymization, image quality, protected patients, or scan availability. Radiologist score of 0, 1, or 2 indicates no lesion, a benign nodule, or indeterminate nodule in the tumor-bearing lobe, respectively
Uncertainty estimation
To gauge algorithm uncertainty, we first calculated the malignancy risk estimation score for all nodules in DLCST and the clinical dataset using a previously established DL-based ensemble algorithm [6]. This DL algorithm predicts a lung nodule malignancy risk score using an ensemble of 10 two-dimensional and 10 three-dimensional convolutional neural networks. The DL algorithm was trained and internally validated on data from NLST. Further details of the training procedure are described in Venkadesh et al (Radiology, 2020) [6]. Data from DLCST and the clinical dataset were not part of the training data of the DL algorithm.
To assess the ensemble’s variability and quantify uncertainty, the entropy was calculated for all 20 distinct DL outputs in the ensemble. Entropy provides a quantitative measure of the randomness of the DL outputs that can be used as an estimation of uncertainty for the predicted malignancy risk. Entropy has already shown its benefits in DL for medical image segmentation and grading of breast carcinoma in previous research [18, 19]. High values of entropy indicate that the DL algorithm is equally torn between predicting a benign or malignant outcome, whereas low entropy indicates that the DL algorithm is confident in its prediction. Entropy can therefore serve as an indicator of algorithm uncertainty. For instance, the entropy reaches its peak when all 20 networks make predictions around 0.5, suggesting uncertainty in classifying the malignancy risk of a nodule. In contrast, the entropy reaches its minimum when all predictions are clustered around either 0 or 1. The final uncertainty score was derived by averaging the entropy values from all 20 distinct DL outputs using the formula below:
where \(\overline{H }\) is the mean entropy, \(N\) is the number of distinct DL outputs, and \({p}_{i}\) is the predicted malignancy risk for the positive class.
Development set
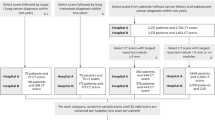
An uncertainty score, using mean entropy, was computed for each nodule in DLCST, resulting in a distribution of uncertainty scores across the entire dataset. Two cut-off values were determined at the 90th and 95th percentiles of this distribution, which were used as discriminative thresholds for classifying nodules into two distinct groups: certain and uncertain. These thresholds resulted in 90% and 95% of the data being retained as certain. This allows us to maintain the largest part of our data in a certain group, hereby mimicking a real-world scenario in which referring a high percentage of cases to human readers would limit and counteract the use of a DL algorithm. Additionally, this allows us to maintain enough data to assess the discriminative performance of the DL algorithm in the uncertain group. A schematic overview of this process is shown in Figure 2.
Schematic overview of how the uncertainty score is utilized to split the dataset into a certain and uncertain group. A nodule block (50x50x50mm) is used as input of the DL algorithm that outputs a malignancy risk and uncertainty score. The uncertainty score is determined based on the score of the individual algorithms in the ensemble. A 90th/95th percentile cut-off value on the uncertainty distribution of all nodules in the dataset is used to split it into a certain and uncertain group to compare algorithm performance. The uncertainty distribution is based on the mean entropy of the individual outputs of the DL algorithm
The discriminative performance of the DL algorithm for a nodule malignancy risk estimation task was compared between the full dataset and certain and uncertain groups using the 90th and 95th percentile thresholds. The discriminative performance of the DL algorithm was assessed by using the area under the receiver operating characteristics curve (AUC) and in terms of sensitivity at 95% specificity. By setting the specificity at 95%, we aimed to minimize false positives. Additionally, a subgroup analysis was conducted on nodule types (solid, part-solid, and non-solid) and nodule size. For nodule size subgroup analysis, nodules were categorized as small (<6mm), medium (≥ 6 to < 8mm), or large (≥8mm), following the size criteria from Lung-RADS 2022 [20]. The subgroup analysis was performed using the 90th percentile threshold to make sure enough nodules remained in each subgroup of the uncertain cases for further analysis. This subgroup examination allows us to gain insights into the DL algorithm’s level of uncertainty within distinct subgroups and unveil nodule characteristics associated with uncertainty.
External validation set
The uncertainty thresholds obtained from the DLCST dataset were applied to nodules of the clinical dataset to investigate whether the DL algorithm exhibits increased uncertainty when dealing with non-screening data. The 90th and 95th percentile thresholds, initially determined on DLCST, were externally validated on the incidentally detected nodules from the clinical dataset. These discriminative thresholds were again used to categorize the nodules into certain and uncertain groups. The performance of the DL algorithm for estimating pulmonary nodule malignancy risk was compared between the full set of nodules in the clinical dataset and certain and uncertain groups using 90th and 95th percentile thresholds of mean entropy.
Furthermore, a subgroup analysis was conducted to discern potential nodule characteristics associated with uncertainty using the 90th percentile threshold of DLCST. This analysis employs the same nodule type and size categories as used for subgroup analysis on the DLCST data. Simultaneously, this approach enabled us to facilitate a comparison between the two datasets, bolstering our understanding of the algorithm's behavior and performance across nodules from distinct study populations.
Statistical analysis
The independent samples t-test was conducted to establish statistical differences in nodule size of certain and uncertain benign and malignant nodules and in nodule size between DLCST and the clinical dataset. The chi-square test was conducted to establish statistical differences in nodule type and size (small, medium, large) of the full dataset and the certain and uncertain groups. For the Chi-square test, part-solid and non-solid types were grouped together as subsolids to account for their small sample size. DeLong’s test was conducted to assess statistical differences between the AUC of the full data and certain and uncertain groups. p values below 0.05 indicated a statistically significant difference.
Results
Participant and nodule characteristics
The DLCST dataset included 599 participants (mean age, 58 years ± 5 [SD]; 484 men) with 883 nodules in 602 CT examinations, and the clinical dataset included 265 participants (mean age, 64 years ± 10 [SD]; 233 men) with 374 nodules in 282 CT examinations. In participants with multiple CT examinations, the first detection of the individual nodules was not observed in the same CT examination. Table 1 shows the patient and nodule characteristics for both datasets. The benign nodules in DLCST were, on average, smaller than the benign nodules in the clinical dataset (p < 0.001). In DLCST and the clinical dataset, 85% and 81% of all benign nodules were solid, respectively (p = 0.305). The malignant nodules in DLCST were, on average, smaller than the malignant nodules in the clinical dataset (p < 0.001). In DLCST, 69% of all malignant nodules were solid, whereas in the clinical dataset, 90% of all malignant nodules were solid (p < 0.001).
Development set
Uncertainty estimation scores were calculated across all 20 predictions of the ensemble algorithm based on mean entropy for all 883 nodules in the DLCST dataset. The entropy-based uncertainty score was 0.51 and 0.59 for the 90th and 95th percentile threshold, respectively. The algorithm achieved an AUC of 0.93 (95% CI: 0.90, 0.96) on the full dataset (Figure 3 and Table 2). With the 90th percentile threshold, the algorithm achieved an AUC of 0.93 (95% CI: 0.88, 0.97) for the certain group and an AUC of 0.62 (95% CI: 0.49, 0.76) for the uncertain group. With the 95th percentile threshold, the algorithm achieved an AUC of 0.94 (95% CI: 0.89, 0.97) for the certain group and an AUC of 0.58 (95% CI: 0.36, 0.79) for the uncertain group. DeLong’s test showed a significant difference in the performance of the DL algorithm between the certain group and the uncertain group (p < 0.001 and p = 0.003) for the 90th and 95th percentile, respectively. No significant differences were found between the full dataset and the certain group for the 90th and 95th percentile (p = 0.935 and p = 0.832, respectively). At 95% specificity, the DL algorithm achieved a sensitivity of 68% on the full dataset, while a sensitivity of 82% was achieved on the certain group of the 90th percentile.
External validation set
By applying the 90th percentile DLCST uncertainty threshold, 27% of the clinical dataset was determined to be uncertain in comparison with 10% of the DLCST dataset. With the 95th percentile DLCST uncertainty threshold, 15% of the clinical dataset was determined to be uncertain in comparison with the 5% on the DLCST dataset.
The DL algorithm achieved an AUC of 0.88 (95% CI: 0.84, 0.91) on the full clinical dataset (Figure 4 and Table 2). By applying the DLCST 90th percentile threshold on the clinical dataset, the algorithm achieved an AUC of 0.90 (95% CI: 0.86, 0.94) for the certain group and an AUC of 0.62 (95% CI: 0.50, 0.73) for the uncertain group. Using the DLCST 95th percentile threshold on the clinical dataset, the algorithm achieved an AUC of 0.89 (95% CI: 0.86, 0.93) for the certain group and an AUC of 0.64 (95% CI: 0.49, 0.78) for the uncertain group. DeLong’s test showed a significant difference in the performance of the DL algorithm between the certain group and the uncertain group of the clinical dataset for the 90th and 95th percentile (p <.001 and p = .004, respectively). No significant differences were found between the full dataset and the certain group for the 90th and 95th percentile (p = .342 and p = .566, respectively). At 95% specificity, the DL algorithm achieved a sensitivity of 32% on the full clinical dataset, while a sensitivity of 34% was achieved on a certain group using the 90th percentile threshold of DLCST. Figures 5 and 6 show examples of nodules where the DL algorithm is certain and uncertain about, together with the malignancy label, DL output, nodule size, and type.
Examples of uncertain cases from the Danish Lung Cancer Screening Trial (DLCST) dataset and the Clinical dataset. Numbers in the bottom right corner of each image indicate the predicted DL malignancy risk, with an extent of color filling in the rings that is proportional to the malignancy risk. A malignancy risk of 0 represents the lowest risk, and 1 represents the highest risk. Arrows indicate the nodule location. DL: Deep Learning Malignancy Risk Estimation. Small: < 6 mm, medium: ≥ 6 to < 8 mm and large: ≥ 8 mm
Examples of certain cases from the Danish Lung Cancer Screening Trial (DLCST) dataset and the Clinical dataset. Numbers in the bottom right corner of each image indicate the predicted DL malignancy risk, with an extent of color filling in the rings that is proportional to the malignancy risk. A malignancy risk of 0 represents the lowest risk, and 1 represents the highest risk. Arrows indicate the nodule location. DL: Deep Learning Malignancy Risk Estimation. Small: < 6 mm, medium: ≥ 6 to < 8 mm, and large: ≥ 8 mm
Subgroup analysis
Table 3 summarizes the characteristics of the certain and uncertain nodules in the DLCST and clinical dataset. In appendix 1 and 2, a complete overview of the number and percentages of nodules considered certain and uncertain per subgroup is given. Nodules were categorized into subgroups based on their size (small, medium, large) and type (solid, part-solid, and non-solid). Analysis of the mean nodule size (mm) shows that in DLCST, the uncertain group consisted of significantly larger benign nodules than the certain group (p < 0.001). In the clinical dataset, the uncertain group also consisted of significantly larger benign nodules than the certain group (p = 0.02) and significantly smaller malignant nodules than the certain group (p < 0.001).
Specifically focusing on the size-based subgroups, the DL algorithm demonstrated increased uncertainty when dealing with medium and large benign nodules as opposed to small benign nodules for both datasets (p = < 0.001). It also demonstrated increased uncertainty about part-solid and non-solid benign nodules compared to solid benign nodules for both datasets (p = < 0.01). For small solid, part-solid, and non-solid nodules in DLCST, only 1.9%, 0%, and 4.8% were considered uncertain, respectively. For large solid, part-solid and non-solid in DLCST, 18.1%, 62.5%, and 48.7% were considered uncertain, respectively. In the clinical dataset, 11.6% of the small solid nodules were considered uncertain in comparison with 23.9% of the large solid nodules. The clinical dataset did not contain any small part-solid or non-solid nodules. However, 52.2% and 46.2% of the large part-solid and non-solid were considered uncertain, respectively.
Discussion
In this retrospective study, we integrated an uncertainty estimation method into a previously developed DL algorithm for estimating pulmonary nodule malignancy risk. We assessed the algorithm’s performance across two uncertainty thresholds, which were determined on screen-detected nodules from the Danish Lung Cancer Screening Trial (DLCST) and externally validated on incidentally detected nodules in a clinical setting. Using the 90th percentile entropy-based uncertainty estimation, the algorithm achieved an AUC of 0.93 (95% CI: 0.88, 0.97) for the certain group and an AUC of 0.62 (95% CI: 0.49, 0.76) for the uncertain group in DLCST. Externally validating this uncertainty threshold on the clinical dataset, the algorithm achieved an AUC of 0.90 (95% CI: 0.86, 0.94) for the certain group and an AUC of 0.62 (95% CI: 0.50, 0.73) for the uncertain group. Further analysis shows that benign nodules in the uncertain group were significantly larger when compared to the certain group, while only in the clinical dataset did malignant nodules had a significantly smaller size. In addition, there was a higher percentage of subsolid nodules in the uncertain group compared to the certain group. These findings highlight nodule characteristics that contribute to DL algorithm uncertainty.
Our uncertainty method showcased a significant decline in AUC for the uncertain groups, underscoring the capacity to identify cases where the DL algorithm harbors doubt in its predictions. These findings align with prior research that ensemble-based DL algorithms provide noteworthy uncertainty assessments by quantifying the prediction variability through approaches such as entropy [11, 14, 21, 22]. However, there is a pivotal difference between these previous studies and ours. We extended beyond the typical uncertainty assessment by providing an examination of nodule characteristics in the uncertain subset. This distinctive approach equips us with the means to not only identify uncertainty but also understand the factors contributing to it as we have seen in our subgroup analysis.
Our subgroup analysis revealed that the uncertain group comprised a higher percentage of larger benign nodules, alongside subsolid nodules. Remarkably, the total proportion of nodules classified as uncertain was 27% in the clinical dataset, whereas this figure was 10% for the screening data from DLCST. This discrepancy shows the elevated levels of uncertainty observed when the DL algorithm encounters nodules with different characteristics acquired in a hospital setting with diverse CT scan protocols instead of a controlled environment such as a screening trial. This further highlights the pivotal role of uncertainty estimation and the importance of validating DL algorithms in target cohort data that were not included in the training phase.
Our work has limitations. The first limitation is that only two uncertainty thresholds were applied. These thresholds were arbitrarily chosen to make sure the largest part of the dataset was determined as certain. Hereby, we intended to mimic a real-world scenario in which referring a high percentage of cases to human readers would limit and counteract the use of an DL algorithm. Nevertheless, the optimal threshold depends on the healthcare setting in which the DL algorithm is applied. We recommend that the optimal uncertainty threshold is discussed in a multi-disciplinary approach to achieve the best clinical benefits, but this should be assessed further in future work. Additionally, a reader study could also evaluate the optimal threshold that balances and maximizes the performance of both the DL algorithm and radiologists. The second limitation is that lung cancer diagnosis in the clinical dataset was obtained through the National Cancer Registry, to which we had to link based on lobe location and incidence date to find the nodule corresponding to the reported cancer. This could have contributed to the selection of more obvious malignancies that are larger in diameter since only nodules with high confidence of representing lung cancer were included.
The quantification of algorithm uncertainty holds valuable potential implications for both clinical practice and model development, as this allows these ambiguous scenarios to be addressed in a more informed and strategic manner. The demonstrated value of uncertainty estimation could help in the clinical implementation of DL algorithms, as this currently remains limited [23, 24]. Especially with the growing detection rate of pulmonary nodules due to an increase in CT examinations [25] and growing interest in lung cancer screening [2, 5, 26], uncertainty estimation could further amplify the benefits DL algorithms have on the anticipated demands of human readers. However, future work should investigate the implications of incorporating such uncertainty estimation within the workflow and performance of human readers using a reader study. Furthermore, additional external validation using newer multicenter data and evaluation among various algorithms is essential to solidify the applicability of this uncertainty estimation. With a similar approach as Alves et al (Radiology, 2023) [16], future research should extend this uncertainty estimation across multiple algorithms, imaging modalities, and cancer types.
In conclusion, we successfully integrated an uncertainty estimation method into a previously developed DL algorithm for nodule malignancy risk estimation and demonstrated that we can identify uncertain cases for which a notable decline in algorithm performance is observed. The nodule characteristics observed in the uncertain group emphasize the prevalence of larger benign nodules and sub-solid nodules, highlighting various factors that contribute to algorithm uncertainty. Integrating uncertainty estimation can be a promising method of mitigating errors, offering clinicians a vital tool to discern cases where expert consultation is needed. In addition, the characteristics of the uncertain cases provide valuable feedback for algorithm developers, highlighting areas where the training data for the algorithm can be extended to improve algorithm performance.
Abbreviations
- AUC:
-
area under the receiver operating characteristics curve
- DL:
-
Deep Learning
- DLCST:
-
Danish Lung Cancer Screening Trial
- IKNL:
-
Netherlands Comprehensive Cancer Organization
- NLST:
-
National Lung Screening Trial
References
The National Lung Screening Trial Research Team, Aberle DR, Adams AM et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. https://doi.org/10.1056/NEJMoa1102873
de Koning HJ, van der Aalst CM, de Jong PA et al (2020) Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med. https://doi.org/10.1056/NEJMoa1911793
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin. https://doi.org/10.3322/caac.21708
Limb M (2022) Shortages of radiology and oncology staff putting cancer patients at risk, college warns. BMJ. https://doi.org/10.1136/bmj.o1430
Wille MM, Dirksen A, Ashraf H et al (2016) Results of the randomized Danish lung cancer screening trial with focus on high-risk profiling. Am J Respir Crit Care Med. https://doi.org/10.1164/rccm.201505-1040OC
Venkadesh KV, Setio AAA, Schreuder A et al (2021) Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology. https://doi.org/10.1148/radiol.2021204433
Faghani S, Moassefi M, Rouzrokh P et al (2023) Quantifying uncertainty in deep learning of radiologic images. Radiology. https://doi.org/10.1148/radiol.222217
Begoli E, Bhattacharya T, Kusnezov D (2019) The need for uncertainty quantification in machine-assisted medical decision making. Nat Mach Intell. https://doi.org/10.1038/s42256-018-0004-1
Kompa B, Snoek J, Beam AL (2021) Second opinion needed: communicating uncertainty in medical machine learning. NPJ Digit Med. https://doi.org/10.1038/s41746-020-00367-3
Herzog L, Murina E, Dürr O, Wegener S, Sick B (2020) Integrating uncertainty in deep neural networks for MRI based stroke analysis. Med Image Anal. https://doi.org/10.1016/j.media.2020.101790
Yang S, Fevens T. (2021) Uncertainty quantification and estimation in medical image classification. ICANN 2021. Springer, Cham.https://doi.org/10.1007/978-3-030-86365-4_54
Linmans J, Elfwing S, van der Laak J, Litjens G (2023) Predictive uncertainty estimation for out-of-distribution detection in digital pathology. Med Image Anal. https://doi.org/10.1016/j.media.2022.102655
Thagaard J, Hauberg S, van der Vegt B, Ebstrup T, Hansen JD, Dahl AB (2020) Can you trust predictive uncertainty under real dataset shifts in digital pathology? MICCAI 2020. Springer, Cham. https://doi.org/10.1007/978-3-030-59710-8_80
Abdar M, Samami M, Dehghani Mahmoodabad S et al (2021) Uncertainty quantification in skin cancer classification using three-way decision-based Bayesian deep learning. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2021.104418
Kurz A, Hauser K, Mehrtens HA et al (2022) Uncertainty estimation in medical image classification: systematic review. JMIR Med Inform. https://doi.org/10.2196/36427
Alves N, Bosma JS, Venkadesh KV et al (2023) Prediction variability to identify reduced AI performance in cancer diagnosis at MRI and CT. Radiology. https://doi.org/10.1148/radiol.230275
Chung K, Mets OM, Gerke PK et al (2018) Brock malignancy risk calculator for pulmonary nodules: validation outside a lung cancer screening population. Thorax. https://doi.org/10.1136/thoraxjnl-2017-211372
Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T (2019) Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing. https://doi.org/10.1016/j.neucom.2019.01.103
Senousy Z, Abdelsamea MM, Mohamed MM, Gaber MM (2021) 3E-Net: entropy-based elastic ensemble of deep convolutional neural networks for grading of invasive breast carcinoma histopathological microscopic images. Entropy. https://doi.org/10.3390/e23050620
American College of Radiology (2022) Lung-RADS® v2022. ACR. Available via https://www.acr.org/-/media/ACR/Files/RADS/Lung-RADS/Lung-RADS-2022.pdf. Accessed 25 October 2023
Dolezal JM, Srisuwananukorn A, Karpeyev D et al (2022) Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology. Nat Commun. https://doi.org/10.1038/s41467-022-34025-x
Lakshminarayanan B, Pritzel A, Blundell C (2017) Simple and scalable predictive uncertainty estimation using deep ensembles. Adv Neural Inf Process Syst. https://doi.org/10.48550/arXiv.1612.01474
Strohm L, Hehakaya C, Ranschaert ER, Boon WPC, Moors EHM (2020) Implementation of artificial intelligence (AI) applications in radiology: hindering and facilitating factors. Eur Radiol. https://doi.org/10.1007/s00330-020-06946-y
Wichmann JL, Willemink MJ, De Cecco CN (2020) Artificial intelligence and machine learning in radiology: current state and considerations for routine clinical implementation. Invest Radiol. https://doi.org/10.1097/rli.0000000000000673
Hendrix W, Rutten M, Hendrix N et al (2023) Trends in the incidence of pulmonary nodules in chest computed tomography: 10-year results from two Dutch hospitals. Eur Radiol. https://doi.org/10.1007/s00330-023-09826-3
Aberle DR, Berg CD, Black WC et al (2011) The National Lung Screening Trial: overview and study design. Radiology. https://doi.org/10.1148/radiol.10091808
Acknowledgements
The authors thank the registration team of the Netherlands Comprehensive Cancer Organization (IKNL) for the collection of data for the National Cancer Registry as well as IKNL staff for scientific advice. We thank Kaman Chung for the data acquisition of the clinical dataset.
Funding
This study has received funding from the Dutch Cancer Society (KWF Kankerbestrijding, project number 14113).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Colin Jacobs.
Conflict of interest
The authors declare the following competing interests:
MP receives grants from Canon Medical Systems, Siemens Healthineers; royalties from Mevis Medical Solutions; and payment for lectures from Canon Medical Systems and Siemens Healthineers. The host institution of MP is a minority shareholder in Thirona. He reports no other relationships that are related to the subject matter of the article.
The host institution of CJ receives research grants and royalties from MeVis Medical Solutions, Bremen, Germany, and payment for lectures from Canon Medical Systems. CJ is a collaborator in a public-private research project where Radboudumc collaborates with Philips Medical Systems (Best, the Netherlands). CJ is a member of the Scientific Editorial Board for European Radiology (Imaging Informatics and Artificial Intelligence). He has not taken part in the selection or review processes for this article. He reports no other relationships that are related to the subject matter of the article.
The host institution of HH receives grants from Siemens Healthineers. He reports no other relationships that are related to the subject matter of the article.
RV is supported by an institutional research grant from Siemens Healthineers.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in DLCST.
Written informed consent was waived by the Institutional Review Board for the clinical dataset.
Ethical approval
Institutional Review Board approval was obtained for DLCST.
Institutional Review Board approval was not required for the clinical dataset because of the retrospective design and data pseudonymization.
Study subjects or cohorts overlap
DLCST nodule cohorts have been previously described in Venkadesh et al. Radiology, 2020 (https://doi.org/https://doi.org/10.1148/radiol.2021204433) and a subset of nodules from the clinical dataset are previously described in Chung et al. Thorax, 2018 (https://doi.org/https://doi.org/10.1136/thoraxjnl-2017-211372).
Methodology
-
Retrospective
-
Experimental
-
Multicenter study
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peeters, D., Alves, N., Venkadesh, K.V. et al. Enhancing a deep learning model for pulmonary nodule malignancy risk estimation in chest CT with uncertainty estimation. Eur Radiol (2024). https://doi.org/10.1007/s00330-024-10714-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00330-024-10714-7