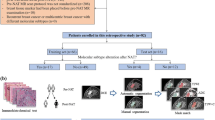
Abstract
Objectives
To investigate whether radiomic features extracted from dynamic susceptibility contrast perfusion-weighted imaging (DSC-PWI) can improve the prediction of the molecular subtypes of adult diffuse gliomas, and to further develop and validate a multimodal radiomic model by integrating radiomic features from conventional and perfusion MRI.
Methods
We extracted 1197 radiomic features from each sequence of conventional MRI and DSC-PWI, respectively. The Boruta algorithm was used for feature selection and combination, and a three-class random forest method was applied to construct the models. We also constructed a combined model by integrating radiomic features and clinical metrics. The models’ diagnostic performance for discriminating the molecular subtypes (IDH wild type [IDHwt], IDH mutant and 1p/19q-noncodeleted [IDHmut-noncodel], and IDH mutant and 1p/19q-codeleted [IDHmut-codel]) was compared using AUCs in the validation set.
Results
We included 272 patients (training set, n = 166; validation set, n = 106) with grade II–IV gliomas (mean age, 48.7 years; range, 19–77 years). The proportions of the molecular subtypes were 66.2% IDHwt, 15.1% IDHmut-noncodel, and 18.8% IDHmut-codel. Nineteen radiomic features (13 from conventional MRI and 6 from DSC-PWI) were selected to build the multimodal radiomic model. In the validation set, the multimodal radiomic model showed better performance than the conventional radiomic model did in predicting the IDHwt and IDHmut-codel subtypes, which was comparable to the conventional radiomic model in predicting the IDHmut-noncodel subtype. The multimodal radiomic model yielded similar performance as the combined model in predicting the three molecular subtypes.
Conclusions
Adding DSC-PWI to conventional MRI can improve molecular subtype prediction in patients with diffuse gliomas.
Key Points
• The multimodal radiomic model outperformed conventional MRI when predicting both the IDH wild type and IDH mutant and 1p/19q-codeleted subtypes of gliomas.
• The multimodal radiomic model showed comparable performance to the combined model in the prediction of the three molecular subtypes.
• Radiomic features from T1-weighted gadolinium contrast-enhanced and relative cerebral blood volume images played an important role in the prediction of molecular subtypes.





Similar content being viewed by others
Abbreviations
- DSC-PWI:
-
Dynamic susceptibility contrast perfusion-weighted imaging
- FISH:
-
Fluorescence in situ hybridization
- FLAIR:
-
Fluid-attenuated inversion recovery
- FOV:
-
Field of view
- IDH:
-
Isocitrate dehydrogenase
- NEX:
-
Number of excite
- rCBF:
-
Relative cerebral blood flow
- rCBV:
-
Relative cerebral blood volume
- T1:
-
T1-weighted
- T1c:
-
T1-weighted gadolinium contrast-enhanced
- T2:
-
T2-weighted
- TE:
-
Echo time
- TR:
-
Repetition time
- VOI:
-
Volume of interest
- WHO:
-
World Health Organization
References
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Stupp R, Hegi ME, Mason WP et al (2009) European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466
Suzuki H, Aoki K, Chiba K et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–468
Louis DN, Perry A, Wesseling P, et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 29:noab106
Cairncross JG, Wang M, Jenkins RB et al (2014) Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol 32:783–790
van den Bent MJ, Brandes AA, Taphoorn MJ et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508
Wong QH, Li KK, Wang WW et al (2021) Molecular landscape of IDH-mutant primary astrocytoma grade IV/glioblastomas. Mod Pathol 34:1245–1260
Cagney DN, Sul J, Huang RY, Ligon KL, Wen PY, Alexander BM (2018) The FDA NIH Biomarkers, EndpointS, and other Tools (BEST) resource in neuro-oncology. Neuro Oncol 20:1162–1172
Rodriguez FJ, Mota RA, Scheithauer BW et al (2009) Interphase cytogenetics for 1p19q and t(1;19)(q10;p10) may distinguish prognostically relevant subgroups in extraventricular neurocytoma. Brain Pathol 19:623–629
Akyerli CB, Yüksel Ş, Can Ö et al (2018) Use of telomerase promoter mutations to mark specific molecular subsets with reciprocal clinical behavior in IDH mutant and IDH wild-type diffuse gliomas. J Neurosurg 128:1102–1114
Yan J, Liu L, Wang W et al (2020) Radiomic features from multi-parameter MRI combined with clinical parameters predict molecular subgroups in patients with medulloblastoma. Front Oncol 10:558162
Coroller TP, Bi WL, Huynh E et al (2017) Radiographic prediction of meningioma grade by semantic and radiomic features. PLoS One 12:e0187908
Zhang B, Chang K, Ramkissoon S et al (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19:109–117
Yan J, Zhang B, Zhang S et al (2021) Quantitative MRI-based radiomics for noninvasively predicting molecular subtypes and survival in glioma patients. NPJ Precis Oncol 5:72
van der Voort SR, Incekara F, Wijnenga MMJ et al (2019) Predicting the 1p/19q codeletion status of presumed low-grade glioma with an externally validated machine learning algorithm. Clin Cancer Res 25:7455–7462
Cui Y, Tha KK, Terasaka S et al (2016) Prognostic imaging biomarkers in glioblastoma: development and independent validation on the basis of multiregion and quantitative analysis of MR images. Radiology 278:546–553
Yan J, Zhang S, Li KK et al (2020) Incremental prognostic value and underlying biological pathways of radiomics patterns in medulloblastoma. EBioMedicine 61:103093
Holdsworth SJ, Bammer R (2008) Magnetic resonance imaging techniques: fMRI, DWI, and PWI. Semin Neurol 28:395–406
Kickingereder P, Sahm F, Radbruch A et al (2015) IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238
Zhang HW, Lyu GW, He WJ et al (2020) DSC and DCE histogram analyses of glioma biomarkers, including IDH, MGMT, and TERT, on differentiation and survival. Acad Radiol 27:e263–e271
Wu H, Tong H, Du X et al (2020) Vascular habitat analysis based on dynamic susceptibility contrast perfusion MRI predicts IDH mutation status and prognosis in high-grade gliomas. Eur Radiol 30:3254–3265
Z wanenburg A, Leger S, V allières M, Löck S (2019) Image biomarker standardisation initiative. Available online at: http://arxiv.org/abs/1612.07003
Breiman L (2001) Random forests. Machine Learn 45:5–32
Louis DN, Perry A, Reifenberger G et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.” Acta Neuropathol 136:805–810
Brat DJ, Aldape K, Colman H et al (2020) cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139:603–608
Zhang X, Tian Q, Wang L et al (2018) Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging 48:916–926
Lu CF, Hsu FT, Hsieh KL et al (2018) Machine learning-based radiomics for molecular subtyping of gliomas. Clin Cancer Res 24:4429–4436
Zhang L, He L, Lugano R et al (2018) IDH mutation status is associated with distinct vascular gene expression signatures in lower-grade gliomas. Neuro Oncol 20:1505–1516
Kapoor GS, Gocke TA, Chawla S et al (2009) Magnetic resonance perfusion-weighted imaging defines angiogenic subtypes of oligodendroglioma according to 1p19q and EGFR status. J Neurooncol 92:373–386
Song S, Wang L, Yang H et al (2021) Static 18F-FET PET and DSC-PWI based on hybrid PET/MR for the prediction of gliomas defined by IDH and 1p/19q status. Eur Radiol 31:4087–4096
Yang X, Lin Y, Xing Z, She D, Su Y, Cao D (2020) Predicting 1p/19q codeletion status using diffusion-, susceptibility-, perfusion-weighted, and conventional MRI in IDH-mutant lower-grade gliomas. Acta Radiol 22:284185120973624
Kim M, Jung SY, Park JE et al (2020) Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur Radiol 30:2142–2151
Yamashita K, Hiwatashi A, Togao O et al (2016) MR imaging-based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. AJNR Am J Neuroradiol 37:58–65
Lee B, Park JE, Bjørnerud A, Kim JH, Lee JY, Kim HS (2018) Clinical value of vascular permeability estimates using dynamic susceptibility contrast MRI: improved diagnostic performance in distinguishing hypervascular primary CNS lymphoma from glioblastoma. AJNR Am J Neuroradiol 39:1415–1422
Macyszyn L, Akbari H, Pisapia JM et al (2016) Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 18:417–425
Funding
This study has received funding from the National Natural Science Foundation of China (No. 82102149, 82273493, 81702465, 82173090, U1804172, U1904148), the Excellent Youth Talent Cultivation Program of Innovation in Health Science and Technology of Henan Province (YXKC2022061), the Key Program of Medical Science and Technique Foundation of Henan Province (No. SBGJ202002062), and the Science and Technology Program of Henan Province (No. 192102310390, 202102310454, 202102310113, 202102310136, 202102310138, 202102310083).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jing Yan.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pei, D., Guan, F., Hong, X. et al. Radiomic features from dynamic susceptibility contrast perfusion-weighted imaging improve the three-class prediction of molecular subtypes in patients with adult diffuse gliomas. Eur Radiol 33, 3455–3466 (2023). https://doi.org/10.1007/s00330-023-09459-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09459-6