Abstract
Objectives
To develop a convolutional neural network system to jointly segment and classify a hepatic lesion selected by user clicks in ultrasound images.
Methods
In total, 4309 anonymized ultrasound images of 3873 patients with hepatic cyst (n = 1214), hemangioma (n = 1220), metastasis (n = 1001), or hepatocellular carcinoma (HCC) (n = 874) were collected and annotated. The images were divided into 3909 training and 400 test images. Our network is composed of one shared encoder and two inference branches used for segmentation and classification and takes the concatenation of an input image and two Euclidean distance maps of foreground and background clicks provided by a user as input. The performance of hepatic lesion segmentation was evaluated based on the Jaccard index (JI), and the performance of classification was based on accuracy, sensitivity, specificity, and the area under the receiver operating characteristic curve (AUROC).
Results
We achieved performance improvements by jointly conducting segmentation and classification. In the segmentation only system, the mean JI was 68.5%. In the classification only system, the accuracy of classifying four types of hepatic lesions was 79.8%. The mean JI and classification accuracy were 68.5% and 82.2%, respectively, for the proposed joint system. The optimal sensitivity and specificity and the AUROC of classifying benign and malignant hepatic lesions of the joint system were 95.0%, 86.0%, and 0.970, respectively. The respective sensitivity, specificity, and the AUROC for classifying four hepatic lesions of the joint system were 86.7%, 89.7%, and 0.947.
Conclusions
The proposed joint system exhibited fair performance compared to segmentation only and classification only systems.
Key Points
• The joint segmentation and classification system using deep learning accurately segmented and classified hepatic lesions selected by user clicks in US examination.
• The joint segmentation and classification system for hepatic lesions in US images exhibited higher performance than segmentation only and classification only systems.
• The joint segmentation and classification system could assist radiologists with minimal experience in US imaging by characterizing hepatic lesions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ultrasound (US) is a widespread first-line imaging modality used in the diagnosis of liver diseases given its low cost, nonionizing characteristics, portable features, and ability for real-time image acquisition and display. Many focal liver lesions are detected incidentally during the first evaluation or follow-up for a primary neoplasm or during surveillance for chronic liver diseases and cirrhosis; however, characterizing the incidental focal hepatic lesions by US imaging is challenging and occasionally shows low sensitivity in the detection of solid lesions due to low contrast between the lesion and the surrounding liver. In addition, large interobserver variability is noted based on the level of operator experience. The confusion created by overlapping US features of hepatic focal lesions is also a factor that limits interpretation [1,2,3]. To overcome these limitations, many computer-aided systems for hepatic lesion segmentation and classification, including deep learning, have been developed [4].
Fully automatic segmentation of existing tumors in US images has been considered difficult due to US imaging limitations, such as speckle noise, low contrast between tumors and surrounding tissues, and varied morphology and echogenicity according to scan direction [5]. In 2016, Xu et al [6] developed a semiautomatic segmentation system based on deep learning, which requires user clicks to segment a specific object in a given image. Despite its original target domain of natural images, this system is also adaptable to clinical images. Deep learning–based systems for lesion classification in US images have also been thoroughly investigated [7,8,9,10,11,12]. Most of the previous approaches take the region-of-interest (ROI), which is manually drawn by a radiologist, as input; however, manual segmentation is tedious and time-consuming, which limits usability in clinical practice [4, 5]. A possible solution involves cascading the segmentation and classification systems, where an ROI automatically extracted by the segmentation system is fed into the classification system. Although the segmentation and classification tasks can be performed separately, they may be related to each other and thus produce mutual benefits when performed simultaneously. Specifically, the segmentation could provide an ROI for classification, and the classification could provide any cues, such as desired shapes based on the lesion types, for segmentation. Therefore, the purpose of this study is to develop a convolutional neural network (CNN) system to jointly segment and classify a hepatic lesion selected by user clicks in US examination.
Materials and methods
This study was performed with approval from our Institutional Review Board. The requirement for informed consent was waived given the retrospective nature of the data analysis and the use of fully anonymized US images.
Dataset development
For developing the dataset, we used 4309 US images with focal hepatic lesions from 3873 patients (1993 men, 1880 women; mean age, 61.0 years ± 11.6; age range, 14–94 years). US examinations were performed between January 2004 and February 2018 at a single institution. In terms of the lesion types contained in the images, 1214 images with hepatic cysts (mean size 16.8 ± 14.9 mm), 1220 images with hemangioma (mean size 17.4 ± 14.5 mm), 1001 images with metastasis (mean size 26.2 ± 15.7 mm), and 874 images with HCC (mean size 23.3 ± 14.7 mm) were included in this study. US data of benign lesions, such as hepatic cysts and hemangiomas, were obtained from patients who were referred to the Department of Radiology at our institution for screening for CLD or other diffuse liver diseases. These cysts and hemangiomas were diagnosed by typical findings in follow-up CT and MRI images. US data of malignant lesions, including metastasis and HCC, were obtained from patients who underwent liver resection or percutaneous liver biopsy for focal hepatic lesions performed within 6 months after US examination. All US images of the focal hepatic lesions regardless of the scanning plane (such as subcostal or intercoastal scan) were collected as grayscale images on the picture archiving and communication system. One representative image with maximum size as measured in the longest dimension of each focal hepatic lesion was used. Manual segmentation of the hepatic lesions was performed by two radiologists (J.Y.L. with 24 years of clinical experience in abdominal US, and H.W.R. with 5 years of clinical experience in abdominal US). These images were used as the reference standard for the lesion segmentation task. For each lesion type, the images were sorted according to the lesion area and divided into 10 groups of equal size (Appendix 1). To include lesions of various sizes in the test set, 10 images were randomly selected from each group for every lesion type, which resulted in 400 images in total. The training set was composed of the remaining 3909 images (Fig. 1). As a constraint, the images of one patient could not be in different sets.
US examinations
US examinations were performed using Siemens Acuson Sequoia, S2000, and S3000 (Siemens AG); Samsung RS 80A ultrasound system (Samsung Medison); LOGIQ E9 ultrasound system (GE Healthcare); and iU22 or EPIQ (Philips Medical systems). The patients underwent US examination after fasting for at least 6 h. Conventional B-mode sonography using a convex probe was performed.
Developing a joint segmentation and classification system
We adapted a semiautomatic segmentation method proposed in [6] for hepatic lesion segmentation. The method takes the concatenation of an input image and two Euclidean distance maps of foreground and background user clicks as inputs and a foreground probability map as shown in Fig. 2a as the output. Therefore, we needed to obtain foreground and background clicks for each image to train the system. Because obtaining these clicks from real users is difficult, we instead used simulated user inputs that are automatically generated by following several rules. For foreground clicks, we randomly selected 1–5 pixels within a lesion (Fig. 3a). For background clicks, 0–10 pixels are randomly selected from the background pixels which are within a certain distance range to the lesion (Fig. 3b). Zero pixels are recorded when a user does not provide any background clicks. Using this method, we generated 15 click sequences for each foreground and background per image. Euclidean distance maps, which represent the minimum Euclidean distance of each pixel to a given set of user clicks, are computed and used as the actual input for the network. An image filled with 255 pixels is used as the background Euclidean distance map if no background click is provided.
Network architecture for the proposed method. a The segmentation only system, which takes a three-channel image composed of a grayscale input image and two Euclidean distance maps of foreground and background clicks provided by a user as input. b The classification only system, which takes a local image patch surrounding a lesion of interest as input. c The joint segmentation and classification system, which takes the same input of the segmentation system and produces segmentation and classification predictions simultaneously. Conv, convolution layer; BN, batch normalization; ReLU, rectified linear unit; FC, fully connected layer; Concat, concatenation
Schematic description of user inputs. The dark gray region represents a lesion of interest, and the shaded region is a set of background pixels that are within a certain distance range to the lesion. a An example of foreground user clicks (blue dots). b An example of background user clicks (red crosses). c An example of bounding box user input (green rectangle), which can be used for testing. d Transformation from the bounding box (c) to user clicks, including one foreground and four background clicks
We used a network architecture (Fig. 2a) adapted from that reported in [13] instead of the fully convolutional networks (FCNs) [14] that were originally used in [6]. The architecture uses bilinear interpolations instead of deconvolution layers for upsampling and has a smaller number of learnable parameters compared to the FCNs, which implies that our network is less prone to overfitting. We added batch normalization layers [15] to our network to further stabilize the network training. The proposed joint segmentation and classification system is implemented by augmenting the segmentation network with an additional classification branch (Fig. 2c). The classification branch, which is composed of several fully connected layers, predicts the lesion type selected by user clicks from the features of the shared convolutional layers. We also implemented the classification only system (Fig. 2b) to compare the performance to that of the joint system to demonstrate any benefit. The system is based on the VGG-16 network [16] with batch normalization [15] and has a structure when the segmentation branch is eliminated from the joint system. We used ideal local image patches centered on a lesion, which are twice the size of the lesion, during training and testing. We used the pixelwise focal loss and α-balanced focal loss for training the segmentation and classification systems, respectively [17]. The sum of two losses was used for the joint system. To train the joint network, we used a stochastic gradient descent optimizer with 0.9 momentum and a weight decay of 5 × 10−4. During the training of 150,000 iterations, the learning rates of 5 × 10−4 and 5 × 10−5 were used for the first and second halves, respectively. For data augmentation, we used horizontal image flipping, image scaling, and random brightness/contrast adjustment. The same hyperparameters were used when training the segmentation only network. For training the classification only network, the learning rates of 10−3 and 10−4 were used for the first and second halves of the whole iterations of 40,000, respectively. For better generalization, the early convolutional layers of all the networks were initialized using the other network trained with breast US images. Because the trained joint system operates with point-type user input, such as clicks, we could have diverse modes in testing. For example, a user scribble can be translated into a set of clicks that could be used for testing. In this study, we used two clinically useful test modes, which are separately based on one click and two clicks. In the one-click mode, a user selects only one pixel within a lesion. On the other hand, a user draws a bounding box surrounding a lesion by placing the top-left and bottom-right corners of it in the two-click mode (Fig. 3c). To test the two-click mode, we again generate five different bounding boxes for each test image by simulation. The top-left and bottom-right corners are sampled with a certain degree of positional error from the tight box of a lesion. The box is enlarged a little to ensure that it includes the lesion. Finally, the bounding box is transformed to five clicks, including one foreground click at the center of it and four background clicks at the corners, when it is used (Fig. 3d).
We used TensorFlow [18] to implement all the networks. The experiments were performed in an environment with Intel Core i7-7700K at 4.2-GHz CPU, 32-GB RAM, and Nvidia GeForce GTX 1080Ti (11 GB VRAM).
Statistical analysis
The segmentation results of the segmentation only system and the joint segmentation and classification system were evaluated against the reference standard and assessed using the Jaccard index (JI) per image. The JI is calculated as the area of the intersection divided by the area of the union between the reference standard and segmentation result [19, 20]. While we present the performance of the two-click mode as our main result, we also provide the results of using a single click for comparison between the two modes. The classification performances of the classification only system and joint system were evaluated using accuracy and the area under the receiver operating characteristic curve (AUC). In terms of the classification, we conducted two experiments to predict benignity and malignancy and categorize the four types of hepatic lesions (hepatic cyst, hemangioma, metastasis, and HCC). We used five different user inputs for each test image to consider the different styles of user input.
Results
Segmentation performance
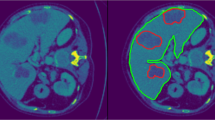
We achieved a mean JI of 68.5% for the segmentation only system in the two-click mode. Compared with this, the joint segmentation and classification system achieved a mean JI of 70.0% when trained with the binary classification task (p < 0.001) and exhibited a comparable result (68.5%) when trained with the four-class classification task (p = 0.95) (Table 1). The comparable performance between the segmentation only system and the joint system was observed regardless of the size and type of hepatic lesions (Appendix 2). Nevertheless, the segmentation results of the joint system showed a tendency to better differentiate the lesion boundaries compared to the segmentation only system (Fig. 4). The mean JI of the joint system markedly decreased to 37.4% in the one-click mode. Although the system still produced accurate segmentations for lesions with clear margins and high contrast, it generated ambiguous and inaccurate boundaries for lesions with unclear margins, large size, and heterogeneous echogenicity (Fig. 5).
Classification performance
The classification accuracies of the classification only system were 89.8% and 79.8% for the binary and four-class classification tasks, respectively. Compared with this, the joint system achieved 90.4% (p = 0.48) and 82.2% (p = 0.01) for the respective tasks (Table 2). The joint system performed favorably against the classification only system across most of the lesion sizes. In terms of the lesion type, the largest increase of accuracy was observed in metastatic lesions (Appendix 3). In terms of ROC analysis, the AUC of the joint system for the binary class classification task was significantly higher than that of the classification only system (0.970, 0.944, p = 0.020). The AUC of the joint system for the four-class classification task was also significantly higher than that of the classification only system (0.947, 0.926, p = 0.040). For the binary classification task, the optimal sensitivity and specificity on the ROC curve of the joint system obtained by calculating Youden’s index [21] were 95.0% and 86.0%, respectively. For the four-class classification, the system yielded a sensitivity of 86.7% and a specificity of 89.7% (Fig. 6, Table 2). The confusion matrix of the four-class classification using the joint system showed that the accuracy for a cyst is the highest, whereas that of HCC is the lowest among all the lesion types (Table 3).
Receiver operating characteristic curves of the classification performance of the classification only system and the joint segmentation and classification system. Two-click user inputs were used for the joint system. a Classification between benign and malignant lesions. b–f Classification of the four types of hepatic lesions, including average (b), cyst (c), hemangioma (Hem.) (d), metastasis (Meta.) (e), and hepatocellular carcinoma (HCC) (f). AUC, area under the receiver operating characteristic curve; Cls-only, classification only system; Joint, joint segmentation and classification system
The mean execution time per image of the joint system was 73 ms with a NVIDIA GeForce GTX 1080Ti GPU card. The respective execution times for the segmentation only system and the classification only system were 73 ms and 10 ms. We note that an image patch should be cropped from a segmentation result before feeding into the classification system, which requires additional processing time, but is not required for the joint system.
Discussion
We proposed a joint segmentation and classification system for hepatic lesions on US images based on user clicks. The system exhibits satisfactory performance in terms of segmentation (mean JI, 70.0% when trained with the binary classification and 68.5% when trained with the four-class classification) and classification (accuracy, 90.4% for the binary classification and 82.2% for the four-class classification). We expected that performing those two tasks jointly would improve the performance of each task compared to solving each task separately. While we decreased the execution time by combining the systems of each task, we also improved the performance for each task.
Hepatic tumor segmentation has long been an important topic, especially in the context of deep learning. Gruber et al developed a sequential network for liver and tumor segmentation in CT images, and a mean JI of 79.2% was reported for tumor segmentation [22]. Vorontsov et al reported per-lesion Dice similarity coefficients of 0.62 – 0.78 for segmentation of colorectal liver metastasis in CT images using a user-correction method. The Dice similarity coefficients were 0.14 – 0.68 without user correction [23]. Compared to the previous studies using CT images, our proposed system exhibited reasonable segmentation performance in US images (mean JI of 68.5 – 70.0%). Interestingly, while the method in [23] showed inaccurate segmentation for small lesions, the proposed system showed reasonable segmentation quality even for small lesions. This finding might be due to inherent differences between US and CT. In this study, we used two kinds of simulated user input, which were one-click and two-click mode. Two-click mode showed better segmentation performance (mean JI of 68.5%) compared with one-click mode (mean JI of 37.4%), especially for lesions with unclear margins, large size, and heterogeneous echogenicity. Considering the role of US in screening for malignancy such as HCC or metastasis, often demonstrating those features, two-click mode seems to be more suitable for clinical practice.
Most previous studies on hepatic tumor segmentation in US images were based on semiautomatic approaches due to the difficulty arising from speckles, shadows, and missing boundaries of US data [5]. Cvancarova et al used the active contour model or snakes to segment hepatic lesions in US images [24, 25]. In this model, segmentation is performed by transforming a snake, which is given by a user, to minimize the energy. When the energy is minimized, balance is achieved between the tension and rigidity of the snake and the degree of fitting to object boundaries. Egger et al proposed a graph-based semiautomatic approach, where the graph is constructed on a circular template placed by a user [26]. In this study, we adapted the semiautomatic segmentation method of [6] for US images, which requires a few user clicks. We used simulated user inputs that reflect different styles of real user input. Learning the user input together with an image enables the network to understand users’ intentions. Because the proposed method performs the segmentation by a trained end-to-end network and does not require complex optimization procedures, it works faster than the previous methods. While the execution times of the previous studies were a few seconds [25, 26], the mean execution time of our system was less than 0.1 s.
Although US is a widely used modality for screening hepatic lesions, it has limitations in terms of diagnostic accuracy of lesions due to its operator-dependent nature, and overlapping sonographic findings of solid hepatic lesions [27]. In clinical practice, US is considered a first-line modality that needs further examinations such as CT or MRI [28]. Human performance of differentiating benign and malignant focal hepatic lesions using only B-mode US images was reported to have high sensitivity (100%), but low specificity (18.9–30.2%) and AUC (0.665–0.706) [29]. Therefore, many studies have been conducted for hepatic lesion classification using deep learning. The system developed by Schmauch et al [30] reported a mean AUC of 0.916 in characterizing five types of hepatic lesions, including hemangioma, metastasis, HCC, cyst, and focal nodular hyperplasia. Hassan et al [31] developed a system classifying four types of focal hepatic lesions, including cyst, hemangioma, HCC, and normal liver parenchyma. The system exhibited good results in terms of accuracy (93.90 – 98.60%), sensitivity (95.70 – 98.30%), and specificity (92.60 – 98.90%). Our proposed system demonstrated higher specificity (86.0%) and AUROC (0.970), but slightly lower sensitivity (95.0%) in differentiating benign and malignant lesions than performance by radiologists in a previous study [29]. These results suggest that our proposed system could be used as a complementary tool for the radiologists. The AUC achieved by our system in characterizing focal hepatic lesions was 0.947, which was higher than the AUC obtained with the classification only system by Schmauch et al [30]. Our proposed system demonstrated slightly lower accuracy (82.2%), sensitivity (86.7%), and specificity (89.7%) compared with the performance of the system developed by Hassan et al [31]; however, we included many metastases cases with heterogeneous echogenicity and ill-defined or irregular borders, which were not included in the previous study by Hassan et al. Both previous studies reported relatively lower accuracy for solid lesions compared with cysts [30, 31]. This tendency is consistent with our results. The reduced accuracy for solid hepatic characterization could be related to overlapping and nonspecific US findings of those tumors. In our study, the classification performance of the joint system was higher than that of the classification only system in both binary and four-class classification tasks. We conjecture that the extracted features for the segmentation task, which is conducted jointly in the system, could help the classification task.
This study has several limitations. Although our dataset includes images obtained using diverse types of US machines, it does not include all different types of machines available in clinical practice; thus, the results may not be applied to US machines not included in this study. In addition, the relatively small numbers of metastases and HCC images in this study might affect the results.
In conclusion, the proposed joint segmentation and classification system for hepatic lesions in US images exhibited higher performance than the segmentation only and classification only systems. The proposed system could assist radiologists with minimal experience in US imaging by characterizing hepatic lesions.
Abbreviations
- AUROC:
-
Area under the receiver operating characteristic curve
- CNN:
-
Convolutional neural network
- FCN:
-
Fully convolutional network
- HCC:
-
Hepatocellular carcinoma
- JI:
-
Jaccard index
- ROI:
-
Region-of-interest
- US:
-
Ultrasound
References
D'Onofrio M, Crosara S, De Robertis R, Canestrini S, Mucelli RP (2015) Contrast-enhanced ultrasound of focal liver lesions. AJR Am J Roentgenol 205:W56–W66
Bommanna Raja K, Madheswaran M, Thyagarajah K (2008) A hybrid fuzzy-neural system for computer-aided diagnosis of ultrasound kidney images using prominent features. J Med Syst 32:65–83
Lee S, Jo IA, Kim KW, Lee JY, Ro YM (2011) Enhanced classification of focal hepatic lesions in ultrasound images using novel texture features2011 18th IEEE International Conference on Image Processing, pp 2025–2028
Park HJ, Park B, Lee SS (2020) Radiomics and deep learning: hepatic applications. Korean J Radiol 21:387–401
Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE (2018) Machine learning for medical ultrasound: status, methods, and future opportunities. Abdom Radiol (NY) 43:786–799
Xu N, Price B, Cohen S, Yang J, Huang TS (2016) Deep interactive object selection Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 373–381
Maclin PS, Dempsey J (1992) Using an artificial neural network to diagnose hepatic masses. J Med Syst 16:215–225
Garra BS, Krasner BH, Horii SC, Ascher S, Mun SK, Zeman RK (1993) Improving the distinction between benign and malignant breast lesions: the value of sonographic texture analysis. Ultrason Imaging 15:267–285
Chan HP, Samala RK, Hadjiiski LM (2020) CAD and AI for breast cancer-recent development and challenges. Br J Radiol 93:20190580
Choi YJ, Baek JH, Park HS et al (2017) A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of thyroid nodules on ultrasound: initial clinical assessment. Thyroid 27:546–552
Hiramatsu Y, Muramatsu C, Kobayashi H, Hara T, Fujita H (2017) Automated detection of masses on whole breast volume ultrasound scanner: false positive reduction using deep convolutional neural network. SPIE
Lekadir K, Galimzianova A, Betriu A et al (2017) A convolutional neural network for automatic characterization of plaque composition in carotid ultrasound. IEEE J Biomed Health Inform 21:48–55
Maninis K-K, Pont-Tuset J, Arbeláez P, Van Gool L (2016) Deep retinal image understanding International conference on medical image computing and computer-assisted intervention. Springer, pp 140–148
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
Lin T-Y, Goyal P, Girshick R, He K, Dollar P (2017) Focal loss for dense object detection 2017 IEEE International Conference on Computer Vision (ICCV), pp 2999–3007
Abadi M, Agarwal A, Barham P et al (2016) Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15:29
van Sloun RJ, Wildeboer RR, Mannaerts CK et al (2019) Deep learning for real-time, automatic, and scanner-adapted prostate (zone) segmentation of transrectal ultrasound, for example, magnetic resonance imaging–transrectal ultrasound fusion prostate biopsy. Eur Urol Focus 7(1):78–85
Youden WJ (1950) Index for rating diagnostic tests. Cancer 3:32–35
Gruber N, Antholzer S, Jaschke W, Kremser C, Haltmeier M (2019) A joint deep learning approach for automated liver and tumor segmentation. arXiv preprint arXiv:190207971
Vorontsov E, Cerny M, Régnier P et al (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiol Artif Intell 1:180014
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1:321–331
Cvancarova M, Albregtsen F, Brabrand K, Samset E (2005) Segmentation of ultrasound images of liver tumors applying snake algorithms and GVFInternational Congress Series. Elsevier, pp 218–223
Egger J, Voglreiter P, Dokter M et al (2016) US-Cut: interactive algorithm for rapid detection and segmentation of liver tumors in ultrasound acquisitions Medical Imaging 2016: Ultrasonic Imaging and Tomography. International Society for Optics and Photonics, pp 97901C
Kim SH, Lee JM, Kim KG et al (2009) Computer-aided image analysis of focal hepatic lesions in ultrasonography: preliminary results. Abdom Imaging 34:183–191
Aubé C, Bazeries P, Lebigot J, Cartier V, Boursier J (2017) Liver fibrosis, cirrhosis, and cirrhosis-related nodules: imaging diagnosis and surveillance. Diagn Interv Imaging 98:455–468
Bartolotta TV, Taibbi A, Galia M et al (2007) Characterization of hypoechoic focal hepatic lesions in patients with fatty liver: diagnostic performance and confidence of contrast-enhanced ultrasound. Eur Radiol 17:650–661
Schmauch B, Herent P, Jehanno P et al (2019) Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging 100:227–233
Hassan TM, Elmogy M, Sallam E-S (2017) Diagnosis of focal liver diseases based on deep learning technique for ultrasound images. Arabian J Sci Eng 42:3127–3140
Funding
This study has received funding from SAMSUNG MEDISON Co., Ltd.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jae Young Lee and Kyoung Mu Lee.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 28 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ryu, H., Shin, S.Y., Lee, J.Y. et al. Joint segmentation and classification of hepatic lesions in ultrasound images using deep learning. Eur Radiol 31, 8733–8742 (2021). https://doi.org/10.1007/s00330-021-07850-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07850-9