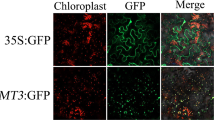
Abstract
Key message
PeTCP10 can be induced by salt stresses and play important regulation roles in salt stresses response in transgenic Arabidopsis.
Abstract
Salt stress is one of the major adverse environmental factors that affect normal plant development and growth. PeTCP10, a Class I TCP member, was markedly expressed in moso bamboo mature leaf, root and stem under normal conditions and also induced by salt stress. Overexpressed PeTCP10 was found to enhance salt tolerance of transgenic Arabidopsis at the vegetative growth stage. It was also found capable to increase relative water content, while decreasing relative electrolyte leakage and Na+ accumulation of transgenic Arabidopsis versus wild-type (WT) plants at high-salt conditions. In addition, it improved antioxidant capacity of transgenic Arabidopsis plants by promoting catalase activity and enhanced their H2O2 tolerance. In contrast to WT plants, transcriptome analysis demonstrated that multiple genes related to abscisic acid, salt and H2O2 response were induced after NaCl treatment in transgenic plants. Meanwhile, overexpressed PeTCP10 improved the tolerance of abscisic acid. Moreover, luciferase reporter assay results showed that PeTCP10 is able to directly activate the expression of BT2 in transgenic plants. In contrary, the germination rates of transgenic plants were significantly lower than those of WT plants under high-NaCl conditions. Both primary root length and survival rate at the seedling stage are also found lower in transgenic plants than in WT plants. It is concluded that overexpressed PeTCP10 enhances salt stress tolerance of transgenic plants at the vegetative growth stage, and it also improves salt sensitiveness in both germination and seedling stages. These research results will contribute to further understand the functions of TCPs in abiotic stress response.










Similar content being viewed by others
Abbreviations
- MDA:
-
Malondialdehyde
- REL:
-
Relative electrical leakage
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- LUC:
-
Luciferase
- CAT:
-
Catalase
- TCP:
-
TEOSINTE BRANCHED 1, CYCLOIDEA and PROLIFERATING CELL FACTOR
- FPKM:
-
Fragments per kilobase of exon model per Million mapped fragments
- RNA-seq:
-
RNA sequencing
- FW:
-
Fresh weight
- SOS:
-
Salt overly sensitive
- Y1H:
-
Yeast one-hybrid
- ABA:
-
Abscisic acid
References
Aguilar-Martínez JA, Sinha N (2013) Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4:406
Almeida DM, Gregorio GB, Oliveira MM, Saibo NJ (2017) Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol Biol 93:61–77
Amin I, Rasool S, Mir MA, Wani W, Masoodi KZ, Ahmad P (2021) Ion homeostasis for salinity tolerance in plants: a molecular approach. Physiol Plant 171:578–594
Arnon DI (1949) Copper enzymes in isolated chloroplasts polyphenoloxidase in beta vulgaris. Plant Physiol 24:1–15
Berriri S, Garcia AV, dit Frey NF, Rozhon W, Pateyron S, Leonhardt N, Montillet JL, Leung J, Hirt H, Colcombet J (2012) Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24:4281–4293
Boudsocq M, Laurière C (2005) Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol 138:1185–1194
Bressan HRA (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Brini F, Masmoudi K (2012) Ion transporters and abiotic stress tolerance in plants. ISRN Mol Biol. https://doi.org/10.5402/2012/927436
Cai R, Dai W, Zhang C, Wang Y, Wu M, Zhao Y, Ma Q, Xiang Y, Cheng B (2017) The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 246:1215–1231
Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143:707–719
Chai W, Jiang P, Huang G, Jiang H, Li X (2017) Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol Mol Biol Plants 23:779–791
Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28:2117–2130
Cheng X, Wang Y, Xiong R, Gao Y, Yan H, Xiang Y (2020) A moso bamboo gene VQ28 confers salt tolerance to transgenic Arabidopsis plants. Planta 251:99
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90:856–867
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18:215–222
Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, Angenent GC, Immink RG (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159:1511–1523
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52
Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protoc. https://doi.org/10.21769/BioProtoc.263
Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24:1923–1928
Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379
Ding S, Cai Z, Du H, Wang H (2019) Genome-wide analysis of TCP family genes in Zea mays L. Identified a role for ZmTCP42 in drought tolerance. Int J Mol Sci. https://doi.org/10.3390/ijms20112762
Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386:485–488
Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, Deng XW (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17:1180–1195
Finlayson SA (2007) Arabidopsis teosinte branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot teosinte branched1. Plant Cell Physiol 48:667–677
Foyer CH, Noctor G (2013) Redox signaling in plants. Antioxid Redox Signal 18:2087–2090
Fu HH, Luan S (1998) AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10:63–73
Gao LW, Yang SL, Wei SW, Huang DF, Zhang YD (2020) Supportive role of the Na+ transporter CmHKT1;1 from Cucumis melo in transgenic Arabidopsis salt tolerance through improved K+/Na+ balance. Plant Mol Biol 103:561–580
Gao Y, Liu H, Zhang K, Li F, Wu M, Xiang Y (2021) A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Rep 40:187–204
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA 114:2419–2424
Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4:1566
Hamel LP, Nicole MC, Duplessis S, Ellis BE (2012) Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell 24:1327–1351
Hou D, Cheng Z, Xie L, Li X, Li J, Mu S, Gao J (2018) The R2R3MYB gene family in Phyllostachys edulis: genome-wide analysis and identification of stress or development-related R2R3MYBs. Front Plant Sci 9:738
Hou D, Zhao Z, Hu Q, Li L, Vasupalli N, Zhuo J, Zeng W, Wu A, Lin X (2020) PeSNAC-1 a NAC transcription factor from moso bamboo (Phyllostachys edulis) confers tolerance to salinity and drought stress in transgenic rice. Tree Physiol 40:1792–1806
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant 6:275–286
Jia F, Wang C, Huang J, Yang G, Wu C, Zheng C (2015) SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J Exp Bot 66:4683–4697
Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69:91–105
Jiu S, Xu Y, Wang J, Wang L, Wang S, Ma C, Guan L, Abdullah M, Zhao M, Xu W, Ma W, Zhang C (2019) Genome-wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera. Front Genet 10:1276
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M (2021) KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49:D545-d551
Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68:147–158
Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9:1607–1619
Krishnamurthy P, Vishal B, Ho WJ, Lok FCJ, Lee FSM, Kumar PP (2020) Regulation of a cytochrome P450 Gene CYP94B1 by WRKY33 transcription factor controls apoplastic barrier formation in roots to confer salt tolerance. Plant Physiol 184:2199–2215
Li D, Zhang Y, Hu X, Shen X, Ma L, Su Z, Wang T, Dong J (2011) Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol 11:109
Li L, Mu S, Cheng Z, Cheng Y, Zhang Y, Miao Y, Hou C, Li X, Gao J (2017) Characterization and expression analysis of the WRKY gene family in moso bamboo. Sci Rep 7:6675
Lian C, Li Q, Yao K, Zhang Y, Meng S, Yin W, Xia X (2018) Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Front Plant Sci 9:954
Liu HL, Wu M, Li F, Gao YM, Chen F, Xiang Y (2018) TCP Transcription Factors in Moso Bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front Plant Sci 9:1263
Liu H, Gao Y, Wu M, Shi Y, Xiang Y (2020a) TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environ Exp Bot. https://doi.org/10.1016/j.envexpbot.2020.104002
Liu S, Yang R, Liu M, Zhang S, Yan K, Yang G, Huang J, Zheng C, Wu C (2020b) PLATZ2 negatively regulates salt tolerance in Arabidopsis seedlings by directly suppressing the expression of the CBL4/SOS3 and CBL10/SCaBP8 genes. J Exp Bot 71:5589–5602
Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98:4782–4787
Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383:794–799
Lykkesfeldt J (2007) Malondialdehyde as biomarker of oxidative damage to lipids caused by smoking. Clin Chim Acta 380:50–58
Ma X, Ma J, Fan D, Li C, Jiang Y, Luo K (2016) Genome-wide identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Sci Rep 6:32795
Mandadi KK, Misra A, Ren S, McKnight TD (2009) BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol 150:1930–1939
Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15:31–39
Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J (2016) Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J Exp Bot 67:3831–3844
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Mukhopadhyay P, Tyagi AK (2015) OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci Rep 5:9998
Munns R, James RA, Läuchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57:1025–1043
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118
Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119:1–11
Pan F, Wu M, Hu W, Liu R, Yan H, Xiang Y (2019) Genome-wide identification and expression analyses of the bZIP transcription factor genes in moso bamboo (Phyllostachys edulis). Int J Mol Sci. https://doi.org/10.3390/ijms20092203
Pandey GK, Kanwar P, Singh A, Steinhorst L, Pandey A, Yadav AK, Tokas I, Sanyal SK, Kim BG, Lee SC, Cheong YH, Kudla J, Luan S (2015) Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol 169:780–792
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, Li Y, Fan D, Guo Y, Li W, Lu Y, Weng Q, Zhou C, Zhang L, Huang T, Zhao Y, Zhu C, Liu X, Yang X, Wang T, Miao K, Zhuang C, Cao X, Tang W, Liu G, Liu Y, Chen J, Liu Z, Yuan L, Liu Z, Huang X, Lu T, Fei B, Ning Z, Han B, Jiang Z (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45:456–461
Ren S, Mandadi KK, Boedeker AL, Rathore KS, McKnight TD (2007) Regulation of telomerase in Arabidopsis by BT2, an apparent target of telomerase activator1. Plant Cell 19:23–31
Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D (2014) Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24:2714–2719
Shan X, Yang K, Xu X, Zhu C, Gao Z (2019) Genome-wide investigation of the NAC gene family and its potential association with the secondary cell wall in moso bamboo. Biomolecules. https://doi.org/10.3390/biom9100609
Smirnoff N (2010) The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol 125:27–58
Song C, Chung WS, Lim CO (2016) Overexpression of heat shock factor gene HsfA3 increases galactinol levels and oxidative stress tolerance in Arabidopsis. Mol Cells 39:477–483
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270
Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. The Plant J 33:513–520
Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63:809–823
Vaahtera L, Brosché M, Wrzaczek M, Kangasjärvi J (2014) Specificity in ROS signaling and transcript signatures. Antioxid Redox Signal 21:1422–1441
Wang ST, Sun XL, Hoshino Y, Yu Y, Jia B, Sun ZW, Sun MZ, Duan XB, Zhu YM (2014) MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE. https://doi.org/10.1371/journal.pone.0091357
Wu M, Liu H, Han G, Cai R, Pan F, Xiang Y (2017) A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci Rep 7:11721
Wu L, Wu M, Liu H, Gao Y, Chen F, Xiang Y (2021) Identification and characterisation of monovalent cation/proton antiporters (CPAs) in Phyllostachys edulis and the functional analysis of PheNHX2 in Arabidopsis thaliana. Plant Physiol Biochem 164:205–221
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217:523–539
Yao X, Ma H, Wang J, Zhang D (2010) Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Integr Plant Biol 49:885–897
Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21:133–139
Zhang P, Wang R, Yang X, Ju Q, Li W, Lü S, Tran LP, Xu J (2020) The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ 43:1925–1943
Zhao Y, Ma Q, Jin X, Peng X, Liu J, Deng L, Yan H, Sheng L, Jiang H, Cheng B (2014) A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol 55:1142–1156
Zhao H, Gao Z, Wang L, Wang J, Wang S, Fei B, Chen C, Shi C, Liu X, Zhang H, Lou Y, Chen L, Sun H, Zhou X, Wang S, Zhang C, Xu H, Li L, Yang Y, Wei Y, Yang W, Gao Q, Yang H, Zhao S, Jiang Z (2018a) Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). GigaScience 7:1–12
Zhao J, Zhai Z, Li Y, Geng S, Song G, Guan J, Jia M, Wang F, Sun G, Feng N, Kong X, Chen L, Mao L, Li A (2018b) Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of Wheat (Triticum aestivum L.). Front PlaNt Sci. https://doi.org/10.3389/fpls.2018.01282
Zhou Y, Zhang D, An J, Yin H, Fang S, Chu J, Zhao Y, Li J (2018) TCP transcription factors regulate shade avoidance via directly mediating the expression of both phytochrome interacting factors and auxin biosynthetic genes. Plant Physiol 176:1850–1861
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31670672) and the 2021 Graduate Innovation Fund of Anhui Agricultural University (Grant No. 2021yjs-12).
Author information
Authors and Affiliations
Contributions
Experiments in this study were designed by XY, XYZ, and LHL. XYZ and LHL performed experiments. GYM and XR performed experimental guidance. WM and ZKM performed data analysis. XYZ drafted the manuscript, LHL provided advice on the manuscript. All authors edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no declarations of interest.
Additional information
Communicated by Ying-Tang Lu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
299_2021_2765_MOESM3_ESM.tif
Supplementary file3 (TIF 54 KB) Sketch map of BT2 upstream promoter regions (a) and expression levels of BT2 after salt stress treatment (b) (RNA-seq data, values of FPKM).
299_2021_2765_MOESM5_ESM.xlsx
Supplementary file5 (XLSX 12 KB) Identification of stress (salt, H2O2 and osmotic stress) responsive DEGs under 200 mM NaCl condition in PeTCP10 OE lines versus WT using RNA-seq method.
Rights and permissions
About this article
Cite this article
Xu, Y., Liu, H., Gao, Y. et al. The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Rep 40, 1971–1987 (2021). https://doi.org/10.1007/s00299-021-02765-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02765-7