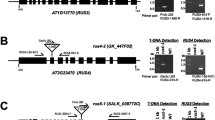
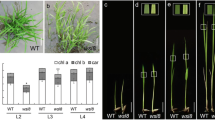
Abstract
Key message
FLA, the homolog of ubiquitin-specific protease does not have deubiquitination activity, but it is essential for flower and chloroplast development in rice.
Abstract
Ubiquitin-specific proteases (UBPs) are widely distributed and highly conserved proteins and are also members of the most important family of deubiquitination enzymes. Although the functions and phylogenies of UBPs from yeast, mammals and Arabidopsis have been widely reported, the functions and evolutionary relationships of UBPs in rice remain unclear. In this study, we characterized the rice flower and leaf color aberrant mutant (fla), which exhibited a variety of developmental defects, including abnormal floral organs and pollen development, and leaf bleaching. We isolated FLA by positional cloning and found that it encodes a homolog of ubiquitin-specific protease. FLA is a ubiquitously expressed gene with the highest expression in floral organs. Subcellular localization analysis indicated that FLA is a cell membrane protein. Through searches of the rice genome database (http://rice.plantbiology.msu.edu), we identified 35 UBP family members in the rice genome. These proteins were grouped into 16 subfamilies based on phylogenetic analysis, and FLA was found to belong to the G8 subfamily. In vitro activity assays revealed that FLA does not have deubiquitination activity. Our data suggest that FLA plays an important role in the development of floral organs and chloroplast in rice, but that this role probably does not involve deubiquitination activity, because FLA does not have an active site and deubiquitination activity.






Similar content being viewed by others
Abbreviations
- UBP/USP:
-
Ubiquitin-specific protease
- DUBs:
-
Deubiquitination enzymes
- MATH:
-
Meprin and TRAF homology
- UBQ:
-
Ubiquitin homolog
- ZnF-UBP:
-
Zinc-finger ubiquitin-specific protease
- UBA:
-
Ubiquitin-associated
- ZnF-MYND:
-
Myeloid, Nervy and DEAF1-type zinc finger
- DUSP:
-
Domain in USPs
- I2–KI:
-
Iodine–potassium iodide
References
Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695:189–207
Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M (1997) In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J 16:4826–4838
Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J (2003) Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep 4:517–522
Belknap WR, Garbarino JE (1996) The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci 1:331–335
Burnett B, Li F, Pittman RN (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet 12:3195–3205
Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S (2013) Deubiquitylases from genes to organism. Physiol Rev 93:1289–1315
Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD (2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27:393–405
Doelling JH, Phillips AR, Soyler-Ogretim G, Wise J, Chandler J, Callis J, Otegui MS, Vierstra RD (2007) The Ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol 145:801–813
Dupre S, Volland C, Haguenauer-Tsapis R (2001) Membrane transport: ubiquitylation in endosomal sorting. Curr Biol 11:R932–R934
Eletr ZM, Wilkinson KD (2014) Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta 1843:114–128
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30:405–439
Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041–1054
Isono E, Nagel MK (2014) Deubiquitylating enzymes and their emerging role in plant biology. Front Plant Sci 5:56
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4
Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10:550–563
Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139:1597–1611
Liu YF, Wang F, Zhang HY, He H, Ma LG, Deng XW (2008) Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J 55:844–856
Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C (2015) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112:11102–11107
Luo M, Luo MZ, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A (2008) UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180:229–236
Moon YK, Hong JP, Cho YC, Yang SJ, An G, Kim WT (2009) Structure and expression of OsUBP6, an Ubiquitin-specific protease 6 homolog in rice (Oryza sativa L.). Mol Cells 28:463–472
Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205
Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786
Pereira RV, Gomes MS, Olmo RP, Souza DM, Cabral FJ, Jannotti-Passos LK, Baba EH, Andreolli ABP, Rodrigues V, Castro-Borges W, Guerra-Sá R (2015) Ubiquitin-specific proteases are differentially expressed throughout the Schistosoma mansoni life cycle. Parasit Vectors 8:349
Pickart CM, Eddins MJ (2004) Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695:55–72
Rao-Naik C, Chandler JS, McArdle B, Callis J (2000) Ubiquitin-specific proteases from Arabidopsis thaliana: cloning of AtUBP5 and analysis of substrate specificity of AtUBP3, AtUBP4, and AtUBP5 using Escherichia coli in vivo and in vitro assays. Arch Biochem Biophys 379:198–208
Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78:363–397
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30
Suresh B, Lee J, Kim KS, Ramakrishna S (2016) The importance of ubiquitination and deubiquitination in cellular reprogramming. Stem Cells Int 2016:6705927
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tank EMH, True HL (2009) Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates. PLoS Genet 5(2):e1000382. https://doi.org/10.1371/journal.pgen.1000382
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8:135–142
Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124:1828–1843
Yu XZ, Sidhu JS, Hong SW, Roinson JF, Ponce RA, Faustman EM (2011) Cadmium induced p53-dependent activation of stress signaling, accumulation of ubiquitinated proteins, and apoptosis in mouse embryonic fibroblast cells. Toxicol Sci 120:403–412
Zhao ZG, Zhu SS, Zhang YH, Bian XY, Wang Y, Jiang L, Chen LM, Liu SJ, Zhang WW, Ikehashi H, Wan JM (2011) Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233:485–495
Zhao J, Zhou H, Zhang M, Gao Y, Li L, Gao Y, Li M, Yang Y, Guo Y, Li X (2016) Ubiquitin-specific protease 24 negatively regulates abscisic acid signaling in Arabidopsis thaliana. Plant Cell Environ 39:427–440
Zhou H, Zhao J, Yang Y, Chen C, Liu Y, Jin X, Chen L, Li X, Deng XW, Schumaker KS, Guo Y (2012) Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24:5106–5122
Zhou J, Wang J, Cheng Y, Chi Y-J, Fan B, Yu JQ, Chen ZX (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9(1):e1003196. https://doi.org/10.1371/journal.pgen.1003196
Zhou J, Zhang Y, Qi JX, Chi YJ, Fan BF, Yu JQ, Chen ZX (2014) E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet 10(1):e1004116. https://doi.org/10.1371/journal.pgen.1004116
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31501287), National Key Research and Development Program of China (2016YFD0100101, 2016YFD0100301), National Science and Technology Support Program of China (2015BAD01B01-1), CAAS Science and Technology Innovation Program, National Infrastructure for Crop Germplasm Resources (NICGR2017-01), Protective Program of Crop Germplasm of China (2017NWB 036-01, 2017NWB036-12-2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Youn-Il Park.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

Supplementary Fig. S1
Comparison of plant agronomic traits between WT and fla. a–c Quantification of the seed set of the main panicle (a), tiller number (b) and height (c) of WT and fla plants. The percentage of the seed set in the main panicle of the fla mutant includes normal seed and seed produced by three types of mutant flowers (types 2, 3, and 5; Fig.1a). Values are the means ± SD (n=20). *P<0.01 (t test). d WT and fla panicles. Bar 5 cm (d) (JPG 450 KB)

Supplementary Fig. S2
Phylogenetic analysis of the UBP gene family in Arabidopsis and rice. Bootstrap values are shown at each node (JPG 2062 KB)
Supplementary Fig. S3
Alignment of the 35 rice UBP proteins. Black and gray boxes denote identical and similar amino acids, respectively (PDF 1570 KB)

Supplementary Fig. S4
Analysis of deubiquitination activity in vitro with rice UBQ10 as the substrate. UBP6 was used as a positive control. Anti-ubiquitin antibody was used to detect ubiquitin. Cleaved products are indicated by black arrowheads. IB, immunoblot (JPG 118 KB)

Supplementary Fig. S5
Analysis of soluble and insoluble pUb protein accumulation in WT and fla. A section of a representative Coomassie brilliant blue-stained SDS-PAGE gel was used as a loading control (bottom). SP, soluble pUb proteins; ISP, insoluble pUb proteins (JPG 23 KB)
Rights and permissions
About this article
Cite this article
Ma, X., Zhang, J., Han, B. et al. FLA, which encodes a homolog of UBP, is required for chlorophyll accumulation and development of lemma and palea in rice. Plant Cell Rep 38, 321–331 (2019). https://doi.org/10.1007/s00299-018-2368-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2368-4